













| General Information: |
|
| Name(s) found: |
STP22 /
YCL008C
[SGD]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 385 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endosome
[IDA ESCRT I complex [IDA |
| Biological Process: |
ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway
[IC telomere maintenance [IMP protein targeting to membrane [IMP protein targeting to vacuole [IMP |
| Molecular Function: |
protein binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSANGKISVP EAVVNWLFKV IQPIYNDGRT TFHDSLALLD NFHSLRPRTR VFTHSDGTPQ 60
61 LLLSIYGTIS TGEDGSSPHS IPVIMWVPSM YPVKPPFISI NLENFDMNTI SSSLPIQEYI 120
121 DSNGWIALPI LHCWDPAAMN LIMVVQELMS LLHEPPQDQA PSLPPKPNTQ LQQEQNTPPL 180
181 PPKPKSPHLK PPLPPPPPPQ PASNALDLMD MDNTDISPTN HHEMLQNLQT VVNELYREDV 240
241 DYVADKILTR QTVMQESIAR FHEIIAIDKN HLRAVEQAIE QTMHSLNAQI DVLTANRAKV 300
301 QQFSSTSHVD DEDVNSIAVA KTDGLNQLYN LVAQDYALTD TIECLSRMLH RGTIPLDTFV 360
361 KQGRELARQQ FLVRWHIQRI TSPLS |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
MVB12 SRN2 STP22 |
| View Details | Krogan NJ, et al. (2006) |
|
MVB12 SRN2 STP22 VPS28 |
| View Details | Qiu et al. (2008) |
|
SNF8 SRN2 STP22 VPS28 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
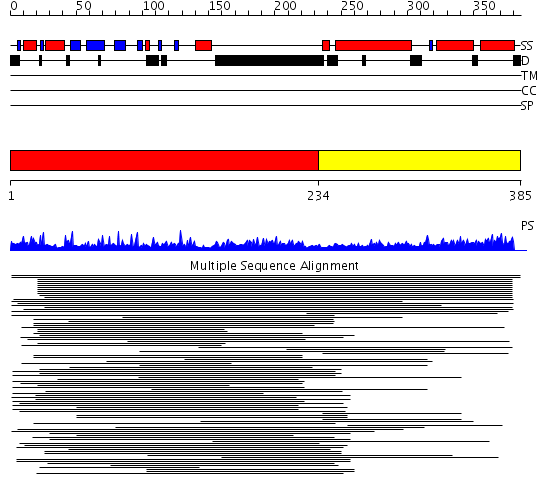
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..233] | 35.69897 | A complex of the Vps23 UEV with ubiquitin | |
| 2 | View Details | [234..385] | 5.119985 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)