













| General Information: |
|
| Name(s) found: |
VPS4 /
YPR173C
[SGD]
|
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 437 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA endoplasmic reticulum [IDA |
| Biological Process: |
protein retention in Golgi apparatus
[IMP late endosome to vacuole transport [IMP sterol metabolic process [IGI |
| Molecular Function: |
ATPase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTGDFLTKG IELVQKAIDL DTATQYEEAY TAYYNGLDYL MLALKYEKNP KSKDLIRAKF 60
61 TEYLNRAEQL KKHLESEEAN AAKKSPSAGS GSNGGNKKIS QEEGEDNGGE DNKKLRGALS 120
121 SAILSEKPNV KWEDVAGLEG AKEALKEAVI LPVKFPHLFK GNRKPTSGIL LYGPPGTGKS 180
181 YLAKAVATEA NSTFFSVSSS DLVSKWMGES EKLVKQLFAM ARENKPSIIF IDEVDALTGT 240
241 RGEGESEASR RIKTELLVQM NGVGNDSQGV LVLGATNIPW QLDSAIRRRF ERRIYIPLPD 300
301 LAARTTMFEI NVGDTPCVLT KEDYRTLGAM TEGYSGSDIA VVVKDALMQP IRKIQSATHF 360
361 KDVSTEDDET RKLTPCSPGD DGAIEMSWTD IEADELKEPD LTIKDFLKAI KSTRPTVNED 420
421 DLLKQEQFTR DFGQEGN |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
SNF7 VPS4 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
SNF7 VPS24 VPS4 |
| View Details | Krogan NJ, et al. (2006) |

|
VPS4 YBL081W |
| View Details | Qiu et al. (2008) |

|
SNF7 VPS4 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #21 Asynchronous Prep2-IMAC Phosphopeptide enrichment A1 | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
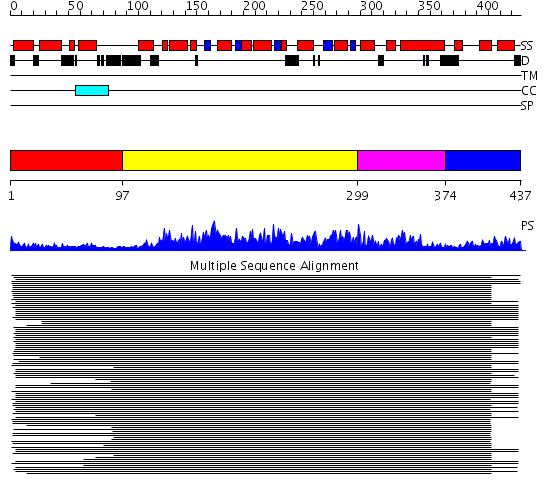
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..96] | 3.600997 | View MSA. Confident ab initio structure predictions are available. | |
| 2 | View Details | [97..298] | 480.0103 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 3 | View Details | [299..373] | 480.0103 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 4 | View Details | [374..437] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)