













| General Information: |
|
| Name(s) found: |
YME1 /
YPR024W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 747 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA i-AAA complex [IDA mitochondrial inner membrane [TAS |
| Biological Process: |
mitochondrion organization
[TAS |
| Molecular Function: |
ATP-dependent peptidase activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNVSKILVSP TVTTNVLRIF APRLPQIGAS LLVQKKWALR SKKFYRFYSE KNSGEMPPKK 60
61 EADSSGKASN KSTISSIDNS QPPPPSNTND KTKQANVAVS HAMLATREQE ANKDLTSPDA 120
121 QAAFYKLLLQ SNYPQYVVSR FETPGIASSP ECMELYMEAL QRIGRHSEAD AVRQNLLTAS 180
181 SAGAVNPSLA SSSSNQSGYH GNFPSMYSPL YGSRKEPLHV VVSESTFTVV SRWVKWLLVF 240
241 GILTYSFSEG FKYITENTTL LKSSEVADKS VDVAKTNVKF DDVCGCDEAR AELEEIVDFL 300
301 KDPTKYESLG GKLPKGVLLT GPPGTGKTLL ARATAGEAGV DFFFMSGSEF DEVYVGVGAK 360
361 RIRDLFAQAR SRAPAIIFID ELDAIGGKRN PKDQAYAKQT LNQLLVELDG FSQTSGIIII 420
421 GATNFPEALD KALTRPGRFD KVVNVDLPDV RGRADILKHH MKKITLADNV DPTIIARGTP 480
481 GLSGAELANL VNQAAVYACQ KNAVSVDMSH FEWAKDKILM GAERKTMVLT DAARKATAFH 540
541 EAGHAIMAKY TNGATPLYKA TILPRGRALG ITFQLPEMDK VDITKRECQA RLDVCMGGKI 600
601 AEELIYGKDN TTSGCGSDLQ SATGTARAMV TQYGMSDDVG PVNLSENWES WSNKIRDIAD 660
661 NEVIELLKDS EERARRLLTK KNVELHRLAQ GLIEYETLDA HEIEQVCKGE KLDKLKTSTN 720
721 TVVEGPDSDE RKDIGDDKPK IPTMLNA |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |

|
MGR1 YME1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
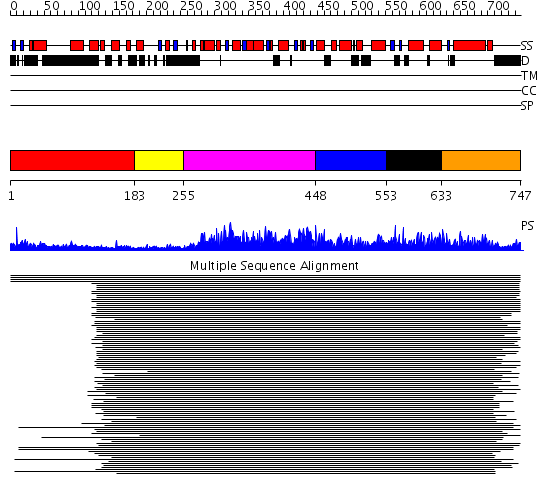
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..182] | 1.577994 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [183..254] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [255..447] | 959.0 | AAA domain of cell division protein FtsH | |
| 4 | View Details | [448..552] | 959.0 | AAA domain of cell division protein FtsH | |
| 5 | View Details | [553..632] | 2.297981 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [633..747] | 6.258988 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)