













| General Information: |
|
| Name(s) found: |
KEL3 /
YPL263C
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 651 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKKNKKDKE AKKARAELKN QKNQKKQEKK FQKNKNKSLN GEEDDESDQD LDEILSSFSK 60
61 KQIELEHVDI TSVEKPSCRT HPLMFANPQH NKHELFIFGG EFTDPETKLT HFYNDLYSYS 120
121 IKNNSWKKYV SQNAPLPRSS AAVAVHPSGI ALLHGGEFSS PKQSKFYHYS DTWLFDCVER 180
181 KFTKLEFGGR DSSPSARSGH RIIAWKNYFI LFGGFRDLGN GQTSYLNDLW CFDISTYKWT 240
241 KLETNSKPDA RSGHCFIPTD NSAILMGGYC KIIAKNNKNL MKGKILNDAW KLNLTPDPKK 300
301 WQWEKLKNFK NQPSPRVGYS FNLWKQNKSV AFGGVYDLQE TEESLESVFY NDLYMFHLEL 360
361 NKWSKLRIKP QRQTNSKNSP ATSKRKSNKD QEKELQDLLN SILAKSNLND DDDDNDDNST 420
421 TGPNSIDDDE DNEDDSDLDN QEDITISNQL PHPRFNAATC VVGDSLFIYS GVWELGEKDY 480
481 PINSFYSIDL NKLDGVKVYW EDLSAIEEAK RLGDRDSDED EFEYEDDEED EDDGEEEQDA 540
541 GPLEGDEDEE SESDDDKQAQ MEIPDERSWL PHPKPFETLR AFYLREGANF LTWSISNNRN 600
601 LKGKQLKTKS FELCEDRWWE RRDQVTLEEE RLEDTGGIIE RDTTTKPSKR R |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
DYN1 KEL3 YRO2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
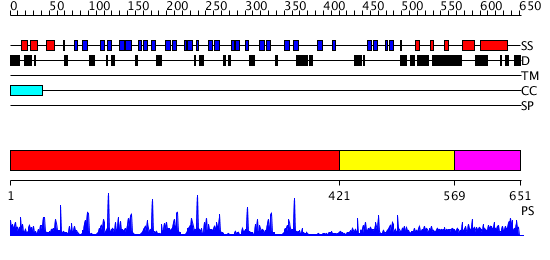
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..420] | 10.0 | NOVEL THIOETHER BOND REVEALED BY A 1.7 ANGSTROMS CRYSTAL STRUCTURE OF GALACTOSE OXIDASE | |
| 2 | View Details | [421..568] | 6.121996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [569..651] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)