













| General Information: |
|
| Name(s) found: |
PDE2 /
YOR360C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 526 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
cAMP-mediated signaling
[IMP |
| Molecular Function: |
3',5'-cyclic-AMP phosphodiesterase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSTLFLIGIH EIEKSQTIVQ NEHYFDRVIE LQDLDSLMVA LYKDRVSPFP NVHNFETGVS 60
61 IVLYDPSKFQ LSVRQLDVLF KRFFPSFNIS AIDHTREENL QRLECVEREN SICRNRITRI 120
121 NHWMYHHHDD TPDGINKNSY GTVNGNSVPT QACEANIYTL LLHLNDSKAQ HLRKASVPRL 180
181 IRNIEFMSFL SDPIEKISQE GSHYWNILST WDFCALSLST QELIWCGFTL IKKLSKDAKV 240
241 LIADNKLLLL LFTLESSYHQ VNKFHNFRHA IDVMQATWRL CTYLLKDNPV QTLLLCMAAI 300
301 GHDVGHPGTN NQLLCNCESE VAQNFKNVSI LENFHRELFQ QLLSEHWPQL LSISKKKFDF 360
361 ISEAILATDM ALHSQYEDRL MHENPMKQIT LISLIIKAAD ISNVTRTLSI SARWAYLITL 420
421 EFNDCALLET FHKAHRPEQD CFGDSYKNVD SPKEDLESIQ NILVNVTDPD DIIKDHPHIP 480
481 NGQIFFINTF AEVFFNALSQ KFSGLKFLSD NVKINKEYWM KHKKPQ |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
COX18 IMD1 IMD2 IMD3 IMD4 PDE2 YAR075W |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #34 Asynchronous SPB prep | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
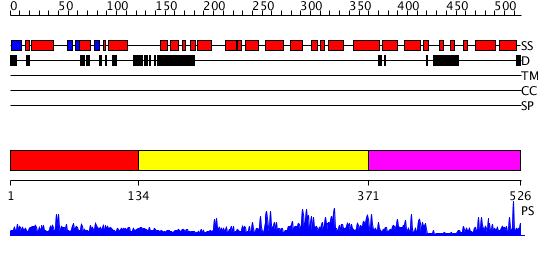
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..133] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [134..370] | 236.0103 | Catalytic domain of cyclic nucleotide phosphodiesterase pde4d | |
| 3 | View Details | [371..526] | 236.0103 | Catalytic domain of cyclic nucleotide phosphodiesterase pde4d |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)