













| General Information: |
|
| Name(s) found: |
MIP1 /
YOR330C
[SGD]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1280 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA |
| Biological Process: |
DNA-dependent DNA replication
[IDA mitochondrial DNA replication [IDA |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDYERTVLKK RSRWGLYVVV EQRGTSMTKL MVRSECMLRM VRRRPLRVQF CARWFSTKKN 60
61 TAEAPRINPV GIQYLGESLQ RQVFGSCGGK DEVEQSDKLM ELSKKSLKDH GLWGKKTLIT 120
121 DPISFPLPPL QGRSLDEHFQ KIGRFNSEPY KSFCEDKFTE MVARPAEWLR KPGWVKYVPG 180
181 MAPVEVAYPD EELVVFDVET LYNVSDYPTL ATALSSTAWY LWCSPFICGG DDPAALIPLN 240
241 TLNKEQVIIG HNVAYDRARV LEEYNFRDSK AFFLDTQSLH IASFGLCSRQ RPMFMKNNKK 300
301 KEAEVESEVH PEISIEDYDD PWLNVSALNS LKDVAKFHCK IDLDKTDRDF FASTDKSTII 360
361 ENFQKLVNYC ATDVTATSQV FDEIFPVFLK KCPHPVSFAG LKSLSKCILP TKLNDWNDYL 420
421 NSSESLYQQS KVQIESKIVQ IIKDIVLLKD KPDFYLKDPW LSQLDWTTKP LRLTKKGVPA 480
481 KCQKLPGFPE WYRQLFPSKD TVEPKITIKS RIIPILFKLS WENSPVIWSK ESGWCFNVPH 540
541 EQVETYKAKN YVLADSVSQE EEEIRTHNLG LQCTGVLFKV PHPNGPTFNC TNLLTKSYNH 600
601 FFEKGVLKSE SELAHQALQI NSSGSYWMSA RERIQSQFVV PSCKFPNEFQ SLSAKSSLNN 660
661 EKTNDLAIII PKIVPMGTIT RRAVENAWLT ASNAKANRIG SELKTQVKAP PGYCFVGADV 720
721 DSEELWIASL VGDSIFNVHG GTAIGWMCLE GTKNEGTDLH TKTAQILGCS RNEAKIFNYG 780
781 RIYGAGAKFA SQLLKRFNPS LTDEETKKIA NKLYENTKGK TKRSKLFKKF WYGGSESILF 840
841 NKLESIAEQE TPKTPVLGCG ITYSLMKKNL RANSFLPSRI NWAIQSSGVD YLHLLCCSME 900
901 YIIKKYNLEA RLCISIHDEI RFLVSEKDKY RAAMALQISN IWTRAMFCQQ MGINELPQNC 960
961 AFFSQVDIDS VIRKEVNMDC ITPSNKTAIP HGEALDINQL LDKSNSKLGK PNLDIDSKVS1020
1021 QYAYNYREPV FEEYNKSYTP EFLKYFLAMQ VQSDKRDVNR LEDEYLRECT SKEYARDGNT1080
1081 AEYSLLDYIK DVEKGKRTKV RIMGSNFLDG TKNAKADQRI RLPVNMPDYP TLHKIANDSA1140
1141 IPEKQLLENR RKKENRIDDE NKKKLTRKKN TTPMERKYKR VYGGRKAFEA FYECANKPLD1200
1201 YTLETEKQFF NIPIDGVIDD VLNDKSNYKK KPSQARTASS SPIRKTAKAV HSKKLPARKS1260
1261 STTNRNLVEL ERDITISREY |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
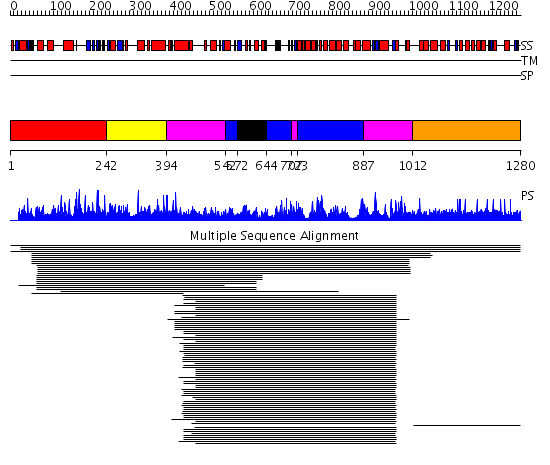
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..241] | 1.013946 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [242..393] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [394..541] [707..722] [887..1011] |
152.0 | Exonuclease domain of prokaryotic DNA polymerase; DNA polymerase I (Klenow fragment) | |
| 4 | View Details | [542..571] [644..706] [723..886] |
152.0 | Exonuclease domain of prokaryotic DNA polymerase; DNA polymerase I (Klenow fragment) | |
| 5 | View Details | [572..643] | 152.0 | Exonuclease domain of prokaryotic DNA polymerase; DNA polymerase I (Klenow fragment) | |
| 6 | View Details | [1012..1280] | 1.007859 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)