













| General Information: |
|
| Name(s) found: |
HAL9 /
YOL089C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1030 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS mitochondrion [IDA |
| Biological Process: |
response to salt stress
[IMP transcription initiation from RNA polymerase II promoter [IMP |
| Molecular Function: |
specific RNA polymerase II transcription factor activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MENQGGDYSP NGFSNSASNM NAVFNNEITG RSDISNVNHQ TGTPRLVPET QIWSMPVPDQ 60
61 LMTMPNRENT LMTGSTIGPN IPMNVAYPNT IYSPTEHQSQ FQTQQNRDIS TMMEHTNSND 120
121 MSGSGKNLKK RVSKACDHCR KRKIRCDEVD QQTKKCSNCI KFQLPCTFKH RDEILKKKRK 180
181 LEIKHHATPG ESLQTSNSIS NPVASSSVPN SGRFELLNGN SPLESNIIDK VSNIQNNLNK 240
241 KMNSKIEKLD RKMSYIIDSV ARLEWLLDKA VKKQEGKYKE KNNLPKPARK IYSTALLTAQ 300
301 KLYWFKQSLG VKASNEEFLS PISEILSISL KWYATQMKKF MDLSSPAFFS SEIILYSLPP 360
361 KKQAKRLLEN FHATLLSSVT GIISLKECLD LAEKYYSESG EKLTYPEHLL LNVCLCSGAS 420
421 ATQSIIRGDS KFLRKDRYDP TSQELKKIEN VALLNAMYYY HKLSTICSGT RTLQALLLLN 480
481 RYFQLTYDTE LANCILGTAI RLAVDMELNR KSSYKSLDFE EAIRRRRMWW HCFCTDKLYS 540
541 LMLSRPPIVG ERDMDMLTDQ NYYEVIKTNI LPDLIDKKED LDKITDVNSA LNVVVNFCQH 600
601 ISLFISYYVS KLVSIESKIY STCFAVRSTL DLSFDAMLDK IKDLNDSLNN WRDNLHVSMK 660
661 LKSYKQYLSV LYAQKSQENP ALSFEIACSR VLNCHFRALY SKVILSMMTT SLLIDNERLY 720
721 KGSRHDIPQL FILFSSQYLN ASKEMLQLFQ GINYQAHMYN EVMYQFSTAM FVLFFYVVDN 780
781 MNDLKKKGEV KEIIDILKKS YDRLVGENDE QLLFDNVKWN TLIVFYSHFL KYVLQRYHAL 840
841 NDSTSIFDSK PYDETITKVI MHSRKIKDET VDQLIMSLKS YGSLHSLQKG NEADLADDGL 900
901 NTNDISSEDF AEEAPINLFG ELSVEILKLL KSHSPISNFG DLSPSSNRKG ISDDSSLYPI 960
961 RSDLTSLVYP IHSSDTGDTL SSGLETPENS NFNSDSGIKE DFEAFRALLP LGKLIYDRDY1020
1021 SFVNTFRDYE |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Toshima J, et al (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
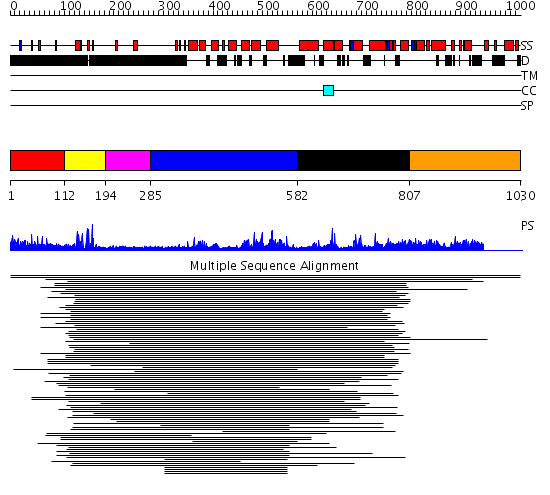
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..111] | 5.178996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [112..193] | 29.37734 | Ethanol regulon transcriptional activator ALCR DNA-binding domain | |
| 3 | View Details | [194..284] | N/A | Confident ab initio structure predictions are available. | |
| 4 | View Details | [285..581] | 6.179981 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [582..806] | 1.083987 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [807..1030] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [849..1030] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)