













| General Information: |
|
| Name(s) found: |
MON2 /
YNL297C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1636 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endosome
[IDA cytosol [TAS extrinsic to membrane [IDA |
| Biological Process: |
endocytosis
[IMP protein targeting to vacuole [IMP transport [IMP |
| Molecular Function: |
guanyl-nucleotide exchange factor activity
[IGI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAMNTGGFDS MQRQLEAELR SLSSESKRRN STIRHASDKS IEILKRVHSF EELERHPDFA 60
61 LPFVLACQSR NAKMTTLAMQ CLQGLSTVPS IPRSRLSEIL DAFIEATHLA MEIQLKVLQV 120
121 VPIFFKTYGK FIYGPLCKKL LLCCSNLLHV PNKAPVVVGT ASATLQQLID EIFDRLSIES 180
181 VVDDKQYEVL ISNSESIKVN VYRYDANKLF DNICSLNEIS SNGAVSDEEM LLDIGDIPID 240
241 YGLEILESIL KNSQKNLLEC QDLQYLLRVK AIPLLLRCIS SSRHFSTAVR SCRCLKLLIR 300
301 KEYLSILELE LEVILSLLIH GISVESNLSG WQRVLSLELF KDLSQDPEIV NTLYMDYDNY 360
361 PDKKHVFKYL LKECIVLLNS PEYITFLAPS KVVEKMDSPL ITTENSTVKT KFMHLLDKSN 420
421 APSINITYII SLILTICNHL CEGLNKSALE SSPLEKKIED KEREEGTGND STVVKVYSGL 480
481 FSGLFELNKL FLYSTSLETS IFHLVVRAFQ KLAHSAGVLS LKDKLRACMK LFSILITNNV 540
541 TSSNQYSFND TSKSAKNQHT RNISTSSVTT SPVESTKNPS RSIADSAQNK EMKRRLHPRN 600
601 ISSRQVSLLR ALISLSISLG PIFDSESWRY TFLTWQWITY YIYGPSADFK ESFYSEDIPP 660
661 PPILTKSDVT SIESSLTKFF ESTSSYSCST FHLVLTRLIL DSKNTLTLEQ TNLNLNNDIG 720
721 YHPLDAKDEI IPCIYNKAFF VNKIGELATY NCKKFLFGKN GKELWSLIST YMIKLISNRE 780
781 MDNDSLRLYT VRVFTDIIKK ATNEVGNSDE QDNKVKQFGT LENLVIDSLM ATINSIKQLD 840
841 IGKQEIYNGT INVESDILFQ LLLTLKEILN EFGELLMNSW TNIFNIINSP FEWTVEDTDF 900
901 SVNEDIDDSS LFEGIVQKHK NMIQVSYDVF KLISDDFLQS LPMSVIKFVI DTLVNFVSQK 960
961 RNLNISFSSI SQFWLVGDYL RVRFNPETLN LSDEKRRSLS EKINNQKLIE IITSSSSHDW1020
1021 ELYNGLWIYL LKNLINCTND DRVEVKNGAV QTFFRIIDSH SVCFPPWDLI FLEVIEPLLT1080
1081 KEWSTEELEN ETDFINVTLQ GLIKLYPEHF KDFKNNTTCA KEWSMLLDFL KRLLSSTSNN1140
1141 TKNAVILNYQ TLLKEIITIE DVPSDILKKC CEIFTDYNIT YSDLSTNASS KTEYDCIYEL1200
1201 ITGFPPLYQL ISKYDAMTDE FVEKVLLLFN SAIKYPLLPE FVQDKTKPSS MQKAILSGLD1260
1261 IFMTNDSKDT EILILLQLST ISILAFDTRE KITKKLGPKL PKASLNRLPT FEAISYMSCS1320
1321 NLRNRIAKID QFGISTLKAK HILRILKNLA EIIKRKSLIT GSESDEIPIW VLASNCFCDL1380
1381 SNKIFKSLQE DAENPLKDNF CDLFINVIVV TLQRINPELD NLTEIDDLNE YSKYREILLE1440
1441 NRIIDLFNER QLDTFIYAVW DCSFLYEFDE LENALMQDCG TFSELSQKLS SFDFSCIFGS1500
1501 TTNPRFQTKY KCSLECLQDL VNFMLNTNEK LRKLTAPYLS ARIALALRRY ISDEYLIGRA1560
1561 PIPKLRKTEL ATLLNGLCVI LRGVLDQNST LGNKQIGVEN LQTLSPLILR TIPVSHKMDG1620
1621 LQDKVLELSL GFTKLD |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
DOP1 MON2 |
| View Details | Gavin AC, et al. (2006) |
|
DOP1 MON2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | 3906: TAP-tagged | Green EM, et al (2005) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
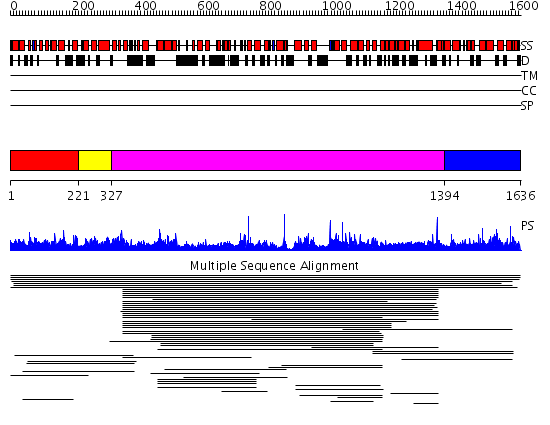
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..220] | 1.012976 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [221..326] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [327..1393] | 8.036826 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [1394..1636] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [663..782] | 1.13 | No description for 2ie3A was found. | |
| 6 | View Details | [783..1559] | 0.974 | AP1 clathrin adaptor core | |
| 7 | View Details | [1560..1636] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)