













| General Information: |
|
| Name(s) found: |
ADE12 /
YNL220W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 433 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
purine nucleotide biosynthetic process
[IMP telomere maintenance [IMP |
| Molecular Function: |
adenylosuccinate synthase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVNVVLGSQW GDEGKGKLVD LLVGKYDIVA RCAGGNNAGH TIVVDGVKYD FHMLPSGLVN 60
61 PNCQNLLGNG VVIHVPSFFK ELETLEAKGL KNARSRLFVS SRAHLVFDFH QVTDKLRELE 120
121 LSGRSKDGKN IGTTGKGIGP TYSTKASRSG LRVHHLVNDQ PGAWEEFVAR YKRLLETRRQ 180
181 RYGDFEYDFE AKLAEYKKLR EQLKPFVVDS VVFMHNAIEA KKKILVEGAN ALMLDIDFGT 240
241 YPYVTSSNTG IGGVLTGLGI PPRTIDEIYG VVKAYTTRVG EGPFPTEQLN ENGEKLQTIG 300
301 AEFGVTTGRK RRCGWLDLVV LKYSTLINGY TSLNITKLDV LDTFKEIPVG ISYSIQGKKL 360
361 DLFPEDLNIL GKVEVEYKVL PGWDQDITKI TKYEDLPENA KKYLKYIEDF VGVPVEWVGT 420
421 GPARESMLHK EIK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
ADE12 TUF1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | 3829 - low filter | Green EM, et al (2005) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
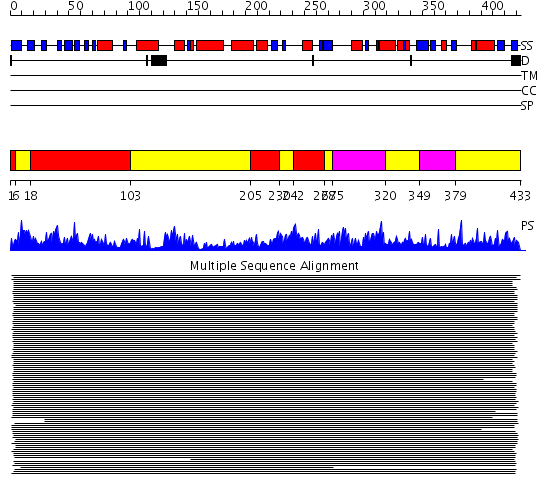
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..5] [18..102] [205..229] [242..267] |
2470.0 | Adenylosuccinate synthetase, PurA | |
| 2 | View Details | [6..17] [103..204] [230..241] [268..274] [320..348] [379..433] |
2470.0 | Adenylosuccinate synthetase, PurA | |
| 3 | View Details | [275..319] [349..378] |
2470.0 | Adenylosuccinate synthetase, PurA |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)