













| General Information: |
|
| Name(s) found: |
SPS19 /
YNL202W
[SGD]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 295 amino acids |
Gene Ontology: |
|
| Cellular Component: |
peroxisomal matrix
[IDA |
| Biological Process: |
ascospore formation
[IEP fatty acid catabolic process [IDA |
| Molecular Function: |
2,4-dienoyl-CoA reductase (NADPH) activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDTMNTANTL DGKFVTEGSW RPDLFKGKVA FVTGGAGTIC RVQTEALVLL GCKAAIVGRD 60
61 QERTEQAAKG ISQLAKDKDA VLAIANVDVR NFEQVENAVK KTVEKFGKID FVIAGAAGNF 120
121 VCDFANLSPN AFKSVVDIDL LGSFNTAKAC LKELKKSKGS ILFVSATFHY YGVPFQGHVG 180
181 AAKAGIDALA KNLAVELGPL GIRSNCIAPG AIDNTEGLKR LAGKKYKEKA LAKIPLQRLG 240
241 STRDIAESTV YIFSPAASYV TGTVLVVDGG MWHLGTYFGH ELYPEALIKS MTSKL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |

|
FOX2 SPS19 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
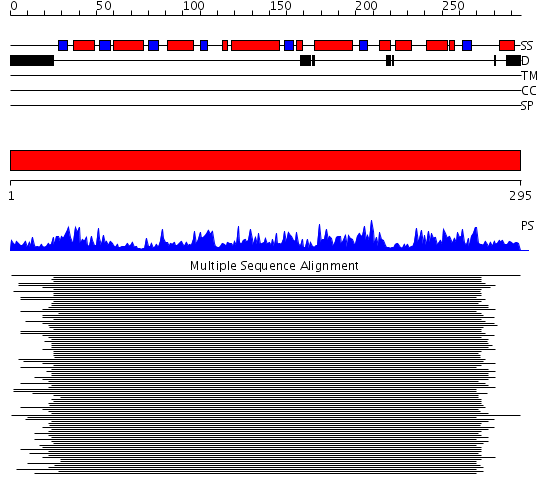
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..295] | 286.383689 | Glucose dehydrogenase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.72 |
Source: Reynolds et al. (2008)