













| General Information: |
|
| Name(s) found: |
ERG12 /
YMR208W
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 443 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[TAS]
|
| Biological Process: |
ergosterol biosynthetic process
[TAS]
|
| Molecular Function: |
mevalonate kinase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLPFLTSAP GKVIIFGEHS AVYNKPAVAA SVSALRTYLL ISESSAPDTI ELDFPDISFN 60
61 HKWSINDFNA ITEDQVNSQK LAKAQQATDG LSQELVSLLD PLLAQLSESF HYHAAFCFLY 120
121 MFVCLCPHAK NIKFSLKSTL PIGAGLGSSA SISVSLALAM AYLGGLIGSN DLEKLSENDK 180
181 HIVNQWAFIG EKCIHGTPSG IDNAVATYGN ALLFEKDSHN GTINTNNFKF LDDFPAIPMI 240
241 LTYTRIPRST KDLVARVRVL VTEKFPEVMK PILDAMGECA LQGLEIMTKL SKCKGTDDEA 300
301 VETNNELYEQ LLELIRINHG LLVSIGVSHP GLELIKNLSD DLRIGSTKLT GAGGGGCSLT 360
361 LLRRDITQEQ IDSFKKKLQD DFSYETFETD LGGTGCCLLS AKNLNKDLKI KSLVFQLFEN 420
421 KTTTKQQIDD LLLPGNTNLP WTS |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
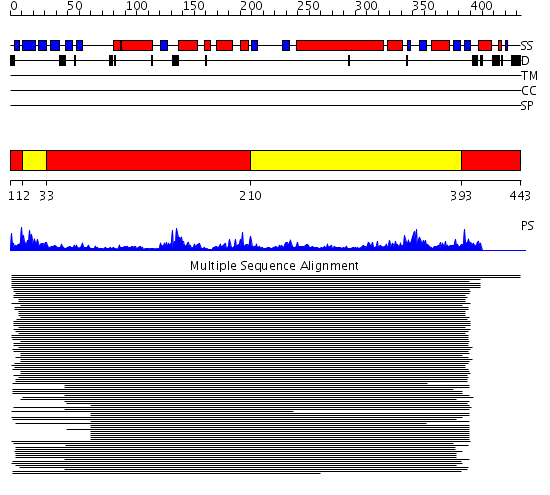
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..11] [33..209] [393..443] |
415.30103 | Mevalonate kinase | |
| 2 | View Details | [12..32] [210..392] |
415.30103 | Mevalonate kinase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|