













| General Information: |
|
| Name(s) found: |
AIM36 /
YMR157C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 255 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLRPLRKSVL ASCRHCFKVC GGLPQKQLPL FSPLLLRARY SSTDSSTKRS NKSDKIDAPG 60
61 FKKIFLVAII GTVIFVKTVQ SLDKNKPKTT LSEEEFENVV KGLKRRVAIF PQGEVDIKFS 120
121 LSPSIEETRK VLQKSQGDDI NELQFVDPVK VIDYYRTLRD DRYEALLNEY YKKYGCDTYA 180
181 YNLPTGMLVM LLGRYFKENF KTGDKLVVVN FPHSIADATR FENEVSIVSK IFVPRKLSGS 240
241 DVCKYYETVG KADII |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |
|
AIM36 MRPL31 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #13 Mitotic Prep2-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
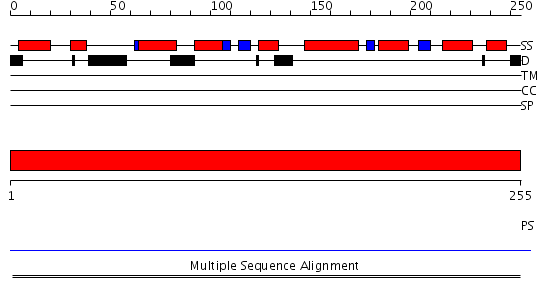
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..255] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [90..255] | 3.26 | SUBSTRATE SPECIFICITY AND ASSEMBLY OF CATALYTIC CENTER DERIVED FROM TWO STRUCTURES OF LIGATED URIDYLATE KINASE |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.66 |
Source: Reynolds et al. (2008)