













| General Information: |
|
| Name(s) found: |
PPR1 /
YLR014C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 904 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[TAS |
| Biological Process: |
regulation of transcription, DNA-dependent
[TAS uracil biosynthetic process [TAS |
| Molecular Function: |
transcription factor activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKQKKFNSKK SNRTDLSKRG DSPNIGISKS RTACKRCRLK KIKCDQEFPS CKRCAKLEVP 60
61 CVSLDPATGK DVPRSYVFFL EDRLAVMMRV LKEYGVDPTK IRGNIPATSD DEPFDLKKYS 120
121 SVSSLGEEGI LPHNGLLADY LVQKGNSMAS SAITSKSMAS PQTINVQRKE FLVNSKKQDG 180
181 SALLPETGSP MTSDARAEEL RRCNKEISAL GTMRESSFNS FLGDSSGISF AKLVFTATNF 240
241 RQDSGDDVLD EDIKQREQKY NGYAEAENNP HFDPLELPPR HAAEVMISRF FVDTNSQLPL 300
301 LHRELFLKKY FEPIYGPWNP NIALASDQTG INSAFEIPIT SAFSAHTEPK RENVTEKIDV 360
361 CSSVDVPWYD TWETSQKVNM RPIVELPTKF HIPYFFLNII FAIGHATQVL KSDITTVATY 420
421 KRRATKYIAS LFSSSDRLEA LAGTLLMVIY SIMRPNVPGV WYTMGSVLRL TVDLGLHSEK 480
481 INKNYDAFTR EIRRRLFWCV YSLDRQICSY FGRPFGIPEE SITTRYPSLL DDSFITLTNR 540
541 EIDDYSDLPS PNPSSKVIAL AMYKIRRIQA SIVRILYAPG AELPRRFMDL ESWRIETYNE 600
601 LERWFQVDVP KNFEMMNCKF NSIWFDLNYH YSKSILYGLS PKFPTLNDTA FKIVLDSTKG 660
661 TIDVFYNLCV NKKIGYTWVA VHNMFMTGMT YLYVNFYSKN NINDCQEKVS EYTEKVLIVL 720
721 KNLIGFCESA KTCYTSYKIL SSVVIKLKFM QINDAKGIFS DSNPLTSQAN RMSSYDKKTN 780
781 VLGFDDGTFD NKVFNRTNFE EKAPFDIPLD EFFTELEKHS NVSQFNTLDV SEGNQVINES 840
841 ASTNTSSALN CQSYTNNQDI MDILFQVTSG SVWDEFFVRS GNGNEGESSY DISKGKNSES 900
901 GGIF |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
BBC1 PPR1 SKT5 UBP7 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
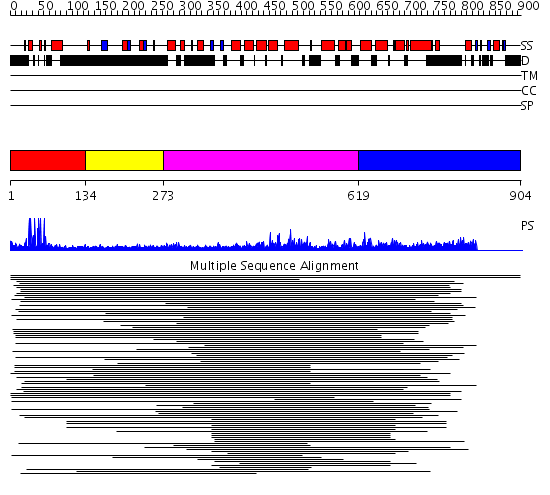
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..133] | 533.751666 | PPR1 | |
| 2 | View Details | [134..272] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [273..618] | 5.104986 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [619..904] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [537..797] | 3.165997 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [798..904] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)