













| General Information: |
|
| Name(s) found: |
YKL075C
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 450 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
response to drug
[IMP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKDLLPKQA ANEPSLKDCT CKRCLKLGAS KEKKIRRKKK GEEKRERHYG NRRKLTFNFL 60
61 KHTNMENTNY DVITSVGYLN EKYGLKKSHY IEKFIKCIHR KINIDVSKIT DAYVNSLNPW 120
121 VKVKLFLLLV TLSEKGGPEY WLDKTDGEKN SEASSTDNSL ENSTKGADSA GSTALRDEMV 180
181 KSHKNLFPTL TEQIIQHNIN QDFTESTYDE DYVFSSIWAN FMEGLINHYL EKVIVPYSEM 240
241 KVCQQLYKPM MKIISLYNEY NELMVKSEKN GFLPSLQDSE NVQGDKGEKE SKDDAVSQER 300
301 LERAQKLLWQ AREDIPKTIS KELTLLSEMY STLSADEQDY ELDEFVCCAE EYIELEYLPA 360
361 LVDVLFANCG TNNFWKIMLV LEPFFYYIED VGGDDDEDED NVDNSEGDEE SLLSRNVEGD 420
421 DNVVERHFKP DPRVITLEKI CEVAARQKWI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
NST1 YKL075C |
| View Details | Krogan NJ, et al. (2006) |
|
DOT6 IBD2 NST1 YKL075C YOL159C-A |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
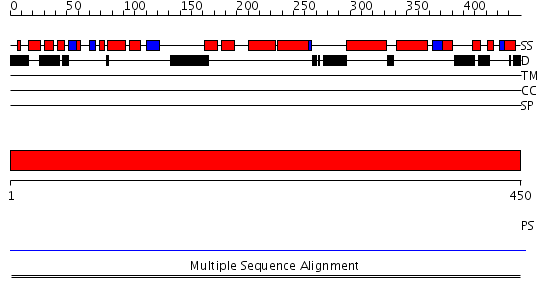
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..450] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [148..285] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [286..450] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)