













| General Information: |
|
| Name(s) found: |
SIP4 /
YJL089W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 829 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
invasive growth in response to glucose limitation
[IMP positive regulation of gluconeogenesis [IMP regulation of transcription from RNA polymerase II promoter [IMP |
| Molecular Function: |
specific RNA polymerase II transcription factor activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKRKYGRSY SLDDTDSCSN KVLIVPTGQS SSNAITDFSV RKAHACDRCR LKKIKCDGLK 60
61 PNCSNCAKID FPCKTSDKLS RRGLPKGYTE LLEKEVVRLT NMNASSSANA NSNLPFINDT 120
121 FYCFDNYNTQ SENQRFLGHL TWNILTNTFP TQKAVVFTDD RNNIDLQLQL LTNFLNLNGD 180
181 FNHLPNFLLL KYDYNLQFLK NLLSVIIKDF FKRQNSLLLL LYPTNLWKNL LLDKINSTAM 240
241 TGEPITLLAL LYIIQFTWSC FDDFKLFKVT KLIVSLTTNS KLDLKVLQLV NLSIFYFMGA 300
301 SVDSCKSKSS LTEHSNVNSV IWTNDLLNLN FTNILNMGLY INPKNLIPIS GNNNNNKSNE 360
361 EDDRIVTFWC FQFLSSWWSL IQGLPKSNFL TEEFQPKSIS VLEIPRLKPF EILLNFIIYS 420
421 LDGCNLLNIS SLNVSDPNFQ FFQNELESFK KNLLLWNLYH NLSDHDNFRF LTSSSNKKLT 480
481 TNLLLKNLTG LNHKLNQPDF VEIQLTLFYL SLKLMTLKEG DQDLKKEDIS LEILSLYFLI 540
541 LTDDSNNDDN QQLQPQQLNL YHFTPFNSID IIDLCLNNLN NWSLSLKYES GQNQPHSSKI 600
601 KFEKFQNFLN HWCPIWYYDE FSTNPFLQIL KINFKLLPFE TIHYSQEEQR LLISLNKLRY 660
661 LDAVSSFNSS SVKSNFASKV NTQLNLLQHS SSNSNFLDAS PYDFNKIFMN NFENYDYETD 720
721 EGYAEDDDEE DSDSDNSLPL EIPFKKSKNK CKNRNKELSQ RLSLFENRDS NSVDFNTDTN 780
781 LNLNPDSPSV TSSKKKYLDH IILDNRDIVS NHDSSKQKFK IQNILNSTF |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
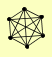
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
GAL83 SIP1 SIP2 SIP4 SNF1 SNF4 |
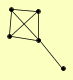
| View Details | Qiu et al. (2008) |
|
CAT8 GAL83 SIP1 SIP2 SIP4 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
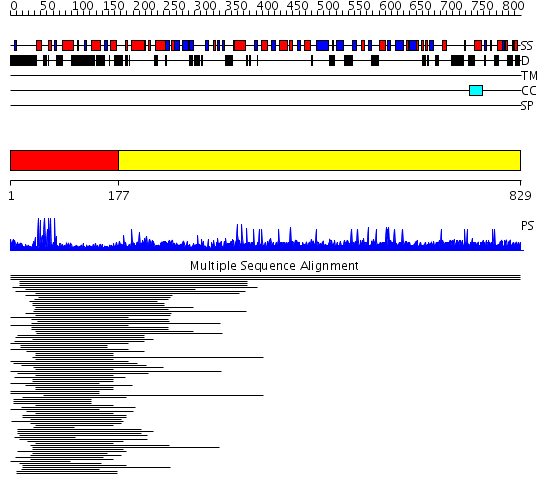
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..176] | 105.30103 | PPR1 | |
| 2 | View Details | [177..829] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)