













| General Information: |
|
| Name(s) found: |
FSP2 /
YJL221C
[SGD]
YIL172C [SGD] |
| Description(s) found:
Found 41 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 589 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA cellular_component [ND] |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
alpha-glucosidase activity
[ISS glucosidase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTISSAHPET EPKWWKEATI YQIYPASFKD SNNDGWGDMK GIASKLEYIK ELGTDAIWIS 60
61 PFYDSPQDDM GYDIANYEKV WPTYGTNEDC FALIEKTHKL GMKFITDLVI NHCSSEHEWF 120
121 KESRSSKTNP KRDWFFWRPP KGYDAEGKPI PPNNWRSYFG GSAWTFDEKT QEFYLRLFCS 180
181 TQPDLNWENE DCRKAIYESA VGYWLDHGVD GFRIDVGSLY SKVAGLPDAP VIDENSKWQL 240
241 SDPFTMNGPR IHEFHQEMNK FIRNRVKDGR EIMTVGEMRH ATDETKRLYT SASRHELSEL 300
301 FNFSHTDVGT SPKFRQNLIP YELKDWKVAL AELFRYVNGT DCWSTIYLEN HDQPRSITRF 360
361 GDDSPKNRVI SGKLLSVLLV SLSGTLYVYQ GQELGEINFK NWPIEKYEDV EVRNNYDAIK 420
421 EEHGENSKEM KRFLEAIALI SRDHARTPMQ WSREEPNAGF SGPNAKPWFY LNESFREGIN 480
481 AEDESKDPNS VLNFWKEALR FRKAHKDITV YGYDFEFIDL DNKKLFSFTK KYDNKTLFAA 540
541 LNFSSDSIDF TIPNNSSSFK LEFGNYPRSE VDASSRTLKP WEGRIYISE |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
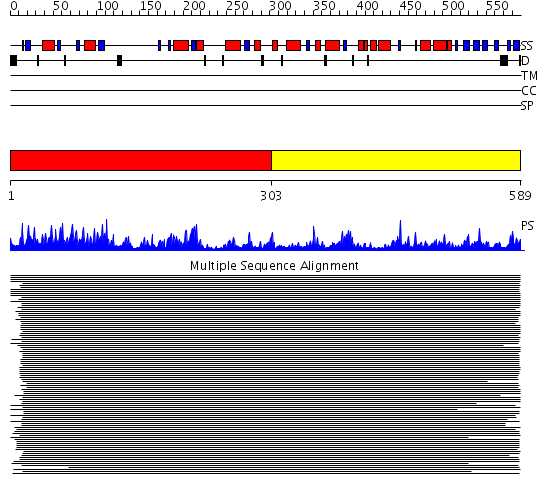
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..302] | 2160.0 | Oligo-1,6-glucosidase; Oligo-1,6, glucosidase | |
| 2 | View Details | [303..589] | 2160.0 | Oligo-1,6-glucosidase; Oligo-1,6, glucosidase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)