













| General Information: |
|
| Name(s) found: |
MTG2 /
YHR168W
[SGD]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 499 amino acids |
Gene Ontology: |
|
| Cellular Component: |
extrinsic to membrane
[IDA mitochondrial inner membrane [IDA |
| Biological Process: |
translation
[IMP |
| Molecular Function: |
GTPase activity
[ISS ribosome binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSIAWSSVFK RELRLERFLP RVYSTKVPDN APRAADNEQW LETLRPITHP EQKKSDHDVS 60
61 YTRHINVPLG EVTSVNYLQR YNKHKHSQGN FVDVRIVKCK SGAGGSGAVS FFRDAGRSIG 120
121 PPDGGDGGAG GSVYIQAVAG LGSLAKMKTT YTAEDGEAGA ARQLDGMRGR DVLIQVPVGT 180
181 VVKWCLPPQK VRELVEREMR KDNNATLRSI LGSTAVNLSV SSGSHRKKIQ LYRHEMAESW 240
241 LFKDKAKEYH ENKDWFKDLH KKMEAYDHSL EQSELFNDQF PLAGLDLNQP MTKPVCLLKG 300
301 GQGGLGNMHF LTNLIRNPRF SKPGRNGLEQ HFLFELKSIA DLGLIGLPNA GKSTILNKIS 360
361 NAKPKIGHWQ FTTLSPTIGT VSLGFGQDVF TVADIPGIIQ GASLDKGMGL EFLRHIERSN 420
421 GWVFVLDLSN KNPLNDLQLL IEEVGTLEKV KTKNILIVCN KVDIDAEKSE SFAKYLQVEK 480
481 FSKSQEWDCV PISALREKT |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
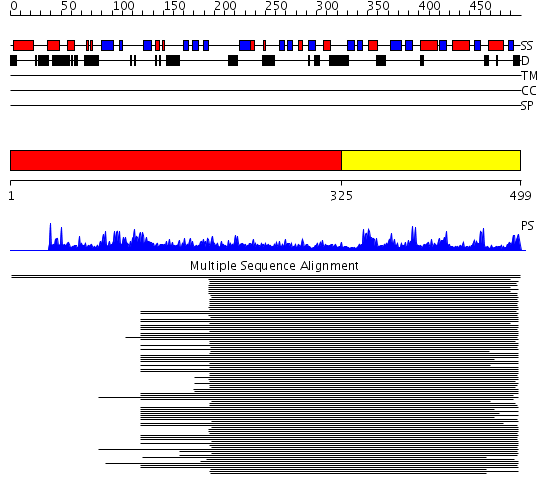
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..324] | 108.29243 | GTP1/OBG No confident structure predictions are available. | |
| 2 | View Details | [325..499] | 62.69897 | ADP-ribosylation factor | |
| 3 | View Details | [128..184] | 13.045757 | Structure of the GTP-binding protein TrmE from Thermotoga maritima | |
| 4 | View Details | [185..499] | 64.0 | Obg GTP-binding protein N-terminal domain; Obg GTP-binding protein C-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)