













| General Information: |
|
| Name(s) found: |
AAP1 /
YHR047C
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 856 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
glycogen metabolic process
[IMP proteolysis [IMP |
| Molecular Function: |
aminopeptidase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSREVLPNNV TPLHYDITLE PNFRAFTFEG SLKIDLQIND HSINSVQINY LEIDFHSARI 60
61 EGVNAIEVNK NENQQKATLV FPNGTFENLG PSAKLEIIFS GILNDQMAGF YRAKYTDKVT 120
121 GETKYMATTQ MEATDARRAF PCFDEPNLKA TFAVTLVSES FLTHLSNMDV RNETIKEGKK 180
181 YTTFNTTPKM STYLVAFIVA DLRYVESNNF RIPVRVYSTP GDEKFGQFAA NLAARTLRFF 240
241 EDTFNIEYPL PKMDMVAVHE FSAGAMENWG LVTYRVIDLL LDIENSSLDR IQRVAEVIQH 300
301 ELAHQWFGNL VTMDWWEGLW LNEGFATWMS WYSCNKFQPE WKVWEQYVTD NLQRALNLDS 360
361 LRSSHPIEVP VNNADEINQI FDAISYSKGS SLLRMISKWL GEETFIKGVS QYLNKFKYGN 420
421 AKTGDLWDAL ADASGKDVCS VMNIWTKRVG FPVLSVKEHK NKITLTQHRY LSTGDVKEEE 480
481 DTTIYPILLA LKDSTGIDNT LVLNEKSATF ELKNEEFFKI NGDQSGIFIT SYSDERWAKL 540
541 SKQANLLSVE DRVGLVADAK ALSASGYTST TNFLNLISNW KNEDSFVVWE QIINSLSALK 600
601 STWVFEPEDI LNALDKFTLD LVLNKLSELG WNIGEDDSFA IQRLKVTLFS AACTSGNEKM 660
661 QSIAVEMFEE YANGNKQAIP ALFKAVVFNT VARLGGENNY EKIFNIYQNP VSSEEKIIAL 720
721 RALGRFEDKE LLERTLSYLL DGTVLNQDFY IPMQGIRVHK KGIERLWAWM QEHWDEIAKR 780
781 LQPGSPVLGG VLTLGLTNFT SFEALEKISA FYSRKVTKGF DQTLAQALDT IRSKAQWVSR 840
841 DREIVATYLR EHEYDQ |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
AAP1 GUD1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
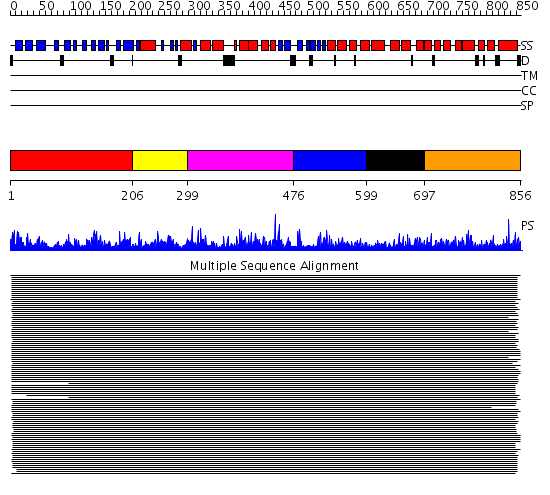
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..205] | 239.06601 | Leukotriene A4 hydrolase C-terminal domain; Leukotriene A4 hydrolase N-terminal domain; Leukotriene A4 hydrolase catalytic domain | |
| 2 | View Details | [206..298] | 239.06601 | Leukotriene A4 hydrolase C-terminal domain; Leukotriene A4 hydrolase N-terminal domain; Leukotriene A4 hydrolase catalytic domain | |
| 3 | View Details | [299..475] | 239.06601 | Leukotriene A4 hydrolase C-terminal domain; Leukotriene A4 hydrolase N-terminal domain; Leukotriene A4 hydrolase catalytic domain | |
| 4 | View Details | [476..598] | 11.39 | Adaptin beta subunit N-terminal fragment | |
| 5 | View Details | [599..696] | 11.39 | Adaptin beta subunit N-terminal fragment | |
| 6 | View Details | [697..856] | 11.39 | Adaptin beta subunit N-terminal fragment |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)