













| General Information: |
|
| Name(s) found: |
DAP2 /
YHR028C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 818 amino acids |
Gene Ontology: |
|
| Cellular Component: |
fungal-type vacuole membrane
[IDA |
| Biological Process: |
protein processing
[ISS |
| Molecular Function: |
dipeptidyl-peptidase and tripeptidyl-peptidase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEGGEEEVER IPDELFDTKK KHLLDKLIRV GIILVLLIWG TVLLLKSIPH HSNTPDYQEP 60
61 NSNYTNDGKL KVSFSVVRNN TFQPKYHELQ WISDNKIESN DLGLYVTFMN DSYVVKSVYD 120
121 DSYNSVLLEG KTFIHNGQNL TVESITASPD LKRLLIRTNS VQNWRHSTFG SYFVYDKSSS 180
181 SFEEIGNEVA LAIWSPNSND IAYVQDNNIY IYSAISKKTI RAVTNDGSSF LFNGKPDWVY 240
241 EEEVFEDDKA AWWSPTGDYL AFLKIDESEV GEFIIPYYVQ DEKDIYPEMR SIKYPKSGTP 300
301 NPHAELWVYS MKDGTSFHPR ISGNKKDGSL LITEVTWVGN GNVLVKTTDR SSDILTVFLI 360
361 DTIAKTSNVV RNESSNGGWW EITHNTLFIP ANETFDRPHN GYVDILPIGG YNHLAYFENS 420
421 NSSHYKTLTE GKWEVVNGPL AFDSMENRLY FISTRKSSTE RHVYYIDLRS PNEIIEVTDT 480
481 SEDGVYDVSF SSGRRFGLLT YKGPKVPYQK IVDFHSRKAE KCDKGNVLGK SLYHLEKNEV 540
541 LTKILEDYAV PRKSFRELNL GKDEFGKDIL VNSYEILPND FDETLSDHYP VFFFAYGGPN 600
601 SQQVVKTFSV GFNEVVASQL NAIVVVVDGR GTGFKGQDFR SLVRDRLGDY EARDQISAAS 660
661 LYGSLTFVDP QKISLFGWSY GGYLTLKTLE KDGGRHFKYG MSVAPVTDWR FYDSVYTERY 720
721 MHTPQENFDG YVESSVHNVT ALAQANRFLL MHGTGDDNVH FQNSLKFLDL LDLNGVENYD 780
781 VHVFPDSDHS IRYHNANVIV FDKLLDWAKR AFDGQFVK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample: hx3 | Hao Xu, et al. (2010) | |
| View Run | Sample: HX11 - both fusion and trans-SNARE complex formation take place. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX13 - trans-SNARE complex forms but fusion is blocked. | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
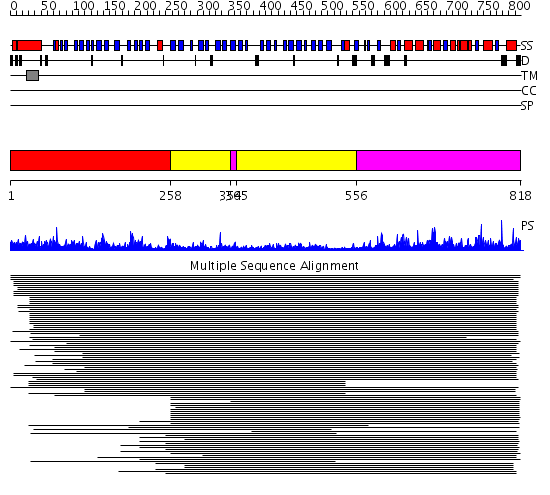
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..257] | 35.09691 | Tup1, C-terminal domain | |
| 2 | View Details | [258..353] [365..555] |
83.0 | Prolyl oligopeptidase, N-terminal domain; Prolyl oligopeptidase, C-terminal domain | |
| 3 | View Details | [354..364] [556..818] |
83.0 | Prolyl oligopeptidase, N-terminal domain; Prolyl oligopeptidase, C-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|