













| General Information: |
|
| Name(s) found: |
IRC7 /
YFR055W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 340 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
cellular copper ion homeostasis
[IEP sulfur metabolic process [ISS |
| Molecular Function: |
cystathionine beta-lyase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIDRTELSKF GITTQLSVIG RNPDEQSGFV NPPLYKGSTI ILKKLSDLEQ RKGRFYGTAG 60
61 SPTIDNLENA WTHLTGGAGT VLSASGLGSI SLALLALSKA GDHILMTDSV YVPTRMLCDG 120
121 LLAKFGVETD YYDPSIGKDI EKLVKPNTTV IFLESPGSGT MEVQDIPALV SVAKKHGIKT 180
181 ILDNTWATPL FFDAHAHGID ISVEAGTKYL GGHSDLLIGL ASANEECWPL LRSTYDAMAM 240
241 LPGAEDCQLA LRGMRTLHLR LKEVERKALD LAAWLGNRDE VEKVLHPAFE DCPGHEYWVR 300
301 DYKGSSGLFS IVLKNGFTRA GLEKMVEGMK VLQLGFSWGG |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
BUD5 IRC7 PHO11 PHO5 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
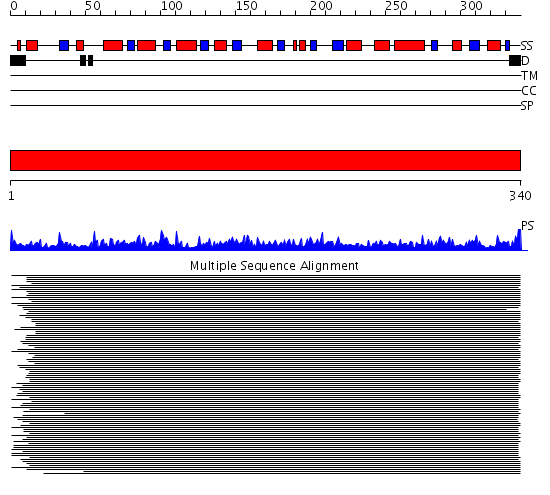
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..340] | 781.30103 | Cystathionine beta-lyase, CBL |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.64 |
Source: Reynolds et al. (2008)