













| General Information: |
|
| Name(s) found: |
EDS1 /
YBR033W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 919 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSHHVPNLYG TPIRDPHERK RNSASMGEVN QSVSSRNCER GSEKGTKQRK KASHACDQCR 60
61 RKRIKCRFDK HTGVCQGCLE VGEKCQFIRV PLKRGPAKKR GSVVSIEKFS SDNDPLQYRP 120
121 RTHSYPMNSG NNYLPSLARN SSFPSISSLF VPSITAQSQQ FVKVPYDDIK RRSSLAILGS 180
181 DSSISTEFGG NYRLDENLNV RQEGKDIVAK GMITPVEEMG ACSSNVRRQG SQSLPIQEQR 240
241 ASPYINPFIS GRSRLSSLSY TSEATTSEGN TQGKNQCMLT PNSVRSIEKE RLNSLTAGFP 300
301 NKKLGTDGRS DKWDKNSTWK PVYRSSNPSH PSTEKNVSLN QEASAKPLML GTYRQFDATS 360
361 FYKVLGIYYN FFHINFPVIP INKSKFTDML DPEKPNVIDE IRQINNEIIQ CFKTALEVLV 420
421 FCKIKQRRSS KSTKSWSRDS LCDFQKGLYY IQNFNKCIAD CFQSLITIKP VLKQNSSVIP 480
481 SRIKFIYFST IIVLNFILIL AGEESSLLLG PSVGVFNEFQ AHKLFLPFQN TSPMLLLNSN 540
541 EESGDEILDY AVLFKRLYIL LNILDTLQSF RLGQPKLINL NFGSAIETYF SDKTGHNQVV 600
601 EKAPVALDNI LRNLKLGEFI TYFVLNRKSL QVNVPHHLLF TNQTDYGEFA VEKGEHDNIA 660
661 GKFETLLKKK EILIRKLLNI EQKNDHILEN CCNSDAEMKN IGELVCSMIT LVSGILDSIT 720
721 NMNAENSVDL DSKPLPNAYF AQDSEEELMS PTQSITSNLA SEENTRCTTK DLMGTVSIFM 780
781 LPMVEECYNI ISLIGPIPTT LISLYIRNGN MAKGINDRIM TLSTALNELV QITALFNTLE 840
841 PFRKNAHDRA KRYYVSATSS TGCYESVMKS MYSGKCAASN ASNVAPSEEE NKKILKKFAD 900
901 IGWKLMDDSE LGCCCCFFN |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman IM, et al. (2002) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
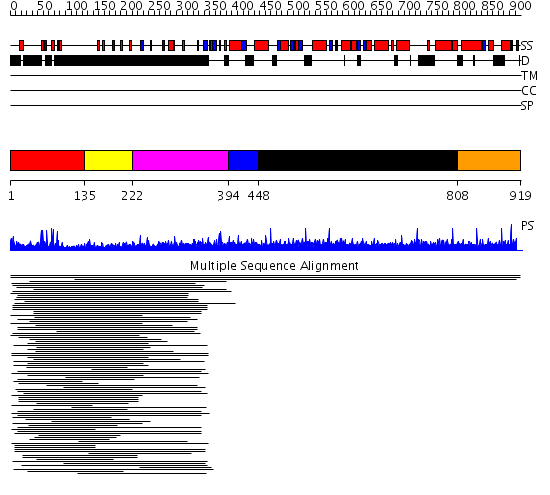
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..134] | 8.69897 | Hap1 (Cyp1); HAP1 | |
| 2 | View Details | [135..221] | 2.255996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [222..393] | 8.233997 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [394..447] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [448..807] | 7.04 | Histidine ammonia-lyase (HAL) | |
| 6 | View Details | [808..919] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 |
|
|||||||||||||||
| 2 | No functions predicted. | |||||||||||||||
| 3 | No functions predicted. | |||||||||||||||
| 4 | No functions predicted. | |||||||||||||||
| 5 | No functions predicted. | |||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.60 |
Source: Reynolds et al. (2008)