













| General Information: |
|
| Name(s) found: |
RKM3 /
YBR030W
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 552 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
phospholipid metabolic process
[IGI |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSVTFKDDVH RILKFVANCN GRFEDSKCDI RESPLGGLGV FAKTDIAEGE SILTLNKSSI 60
61 FSASNSSIAN LLCDSSIDGM LALNIAFIYE TTVFRNSSHW YPFLRTIRIR DDEGHLNLPP 120
121 SFWHADAKRL LKGTSFDTLF DSLAPEEEIM EGFEIAVDLA HKWNDEFGLE IPKGFLDVSE 180
181 ENHEEDYNLK LEKFISVAYT LSSRGFEIDA YHETALVPIA DLFNHHVSDP DLKFVSLYDV 240
241 CDKCGEPDMC KHLIAEEYLE AENLDKNMPK VASMETRVID EDLIKSLEND LEKEYSNVTA 300
301 NIEDDDGGIE NPDECVDLVL KNDVAQGQEI FNSYGELSNV FLLARYGFTV PENQYDIVHL 360
361 GPDFMKILKK EEKYQEKVKW WSQVGHGLFS AWYAQMRQED EEDEDGQAKS DNLSDDIESE 420
421 EEEEEEEGDD SLESWLSQLY IDSSGEPSPS TWALANLLTL TAVQWESLFS KKATPHISDS 480
481 IVNEEKLPFL AKKDNPHSKK LLSNLLKEKQ LPCIKGDNSS KITSATKSML QNARTLVQSE 540
541 HNILDRCLKR LS |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
MAP1 RKM3 |
| View Details | Krogan NJ, et al. (2006) |

|
MAP1 RKM3 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
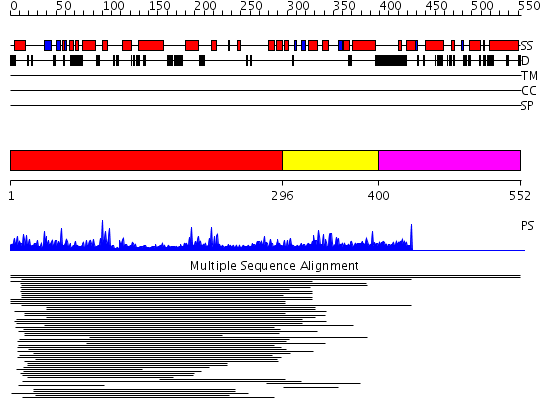
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..295] | 18.046997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [296..399] | 2.028989 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [400..552] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)