













| General Information: |
|
| Name(s) found: |
IMB2_SCHPO
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe 972h- |
| Length: | 910 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGDNPWVLQE QVLVELSEVI KNSLSENSQT RNAALNLLEK AKDIPDLNNY LTCILINATE 60
61 LSVSIRSAAG LLLKNNVRVS SLESGSGLQS LDYTKSTVIR GLCDPEQLIR GISGNVITTI 120
121 ISRWGISTWP EVLPQLMEML SSPASTTQEG AFSALTKICE DSAQELDRDF NGTRPLDFMI 180
181 PRFIELARHE NPKIRTDALF CLNQFVLIQS QSLYAHIDTF LETCYALATD VSPNVRKNVC 240
241 QALVYLLDVR PDKIAPSLGS IVEYMLYSTQ DSDQNVALEA CEFWLAIAEQ PDLCSALGPY 300
301 LDKIVPMLLQ GMVYSDMDLL LLGNDADDYD VEDREEDIRP QHAKGKSRIT LNTQGPITQQ 360
361 GSSNADADEL EDEDEDDDEF DEDDDAFMDW NLRKCSAAAL DVLSSFWKQR LLEIILPHLK 420
421 QSLTSEDWKV QEAGVLAVGA IAEGCMDGMV QYLPELYPYF LSLLDSKKPL VRTITCWTLG 480
481 RYSKWASCLE SEEDRQKYFV PLLQGLLRMV VDNNKKVQEA GCSAFAILEE QAGPSLVPYL 540
541 EPILTNLAFA FQKYQRKNVL ILYDAVQTLA DYVGSALNDK RYIELLITPL LQKWSMIPDD 600
601 DPNLFPLFEC LSSVAVALRD GFAPFAAETY ARTFRILRNT LYLITTAQND PTVDVPDRDF 660
661 LVTTLDLVSG IIQALGSQVS PLLAQADPPL GQIIGICAKD EVPEVRQSAY ALLGDMCMYC 720
721 FDQIRPYCDA LLVDMLPQMQ LPLLHVSASN NAIWSAGEMA LQLGKDMQQW VKPLLERLIC 780
781 ILKSKKSNTT VLENVAITIG RLGVYNPELV APHLELFYQP WFEIIKTVGE NEEKDSAFRG 840
841 FCNILACNPQ ALSYLLPMFV LCVAEYENPS AELRDMFQKI LQGSVELFNG KASWQASPEV 900
901 LAQIQAQYGV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
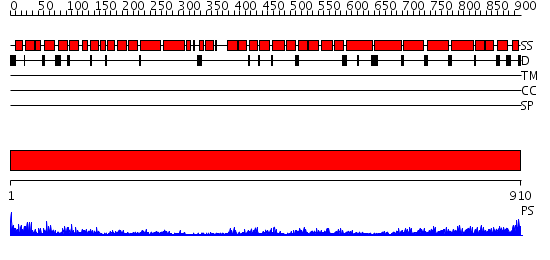
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..910] | 136.0 | Karyopherin beta2 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)