













| General Information: |
|
| Name(s) found: |
gi|83584518
[NCBI NR]
gi|194440160 [NCBI NR] gi|194420903 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli 101-1 |
| Length: | 928 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVQIPQNPLI LVDGSSYLYR AYHAFPPLTN SAGEPTGAMY GVLNMLRSLI MQYKPTHAAV 60
61 VFDAKGKTFR DELFEHYKSH RPPMPDDLRA QIEPLHAMVK AMGLPLLAVS GVEADDVIGT 120
121 LAREAEKAGR PVLISTGDKD MAQLVTPNIT LINTMTNTIL GPEEVVNKYG VPPELIIDFL 180
181 ALMGDSSDNI PGVPGVGEKT AQALLQGLGG LDTLYAEPEK IAGLSFRGAK TMAAKLEQNK 240
241 EVAYLSYQLA TIKTDVELEL TCEQLEVQQP AAEELLGLFK KYEFKRWTAD VEAGKWLQAK 300
301 GAKPAAKPQE TSVADEAPEV TATVISYDNY VTILDEETLK AWIAKLEKAP VFAFDTETDS 360
361 LDNISANLVG LSFAIEPGVA AYIPVAHDYL DAPDQISRER ALELLKPLLE DEKALKVGQN 420
421 LKYDRGILAN YGIELRGIAF DTMLESYILN SVAGRHDMDS LAERWLKHKT ITFEEIAGKG 480
481 KNQLTFNQIA LEEAGRYAAE DADVTLQLHL KMWPDLQKHK GPLNVFENIE MPLVPVLSRI 540
541 ERNGVKIDPK VLHNHSEELT LRLAELEKKA HEIAGEEFNL SSTKQLQTIL FEKQGIKPLK 600
601 KTPGGAPSTS EEVLEELALD YPLPKVILEY RGLAKLKSTY TDKLPLMINP KTGRVHTSYH 660
661 QAVTATGRLS STDPNLQNIP VRNEEGRRIR QAFIAPEDYV IVSADYSQIE LRIMAHLSRD 720
721 KGLLTAFAEG KDIHRATAAE VFGLPLETVT SEQRRSAKAI NFGLIYGMSA FGLARQLNIP 780
781 RKEAQKYMDL YFERYPGVLE YMERTRAQAK EQGYVETLDG RRLYLPDIKS SNGARRAAAE 840
841 RAAINAPMQG TAADIIKRAM IAVDAWLQAE QPRVRMIMQV HDELVFEVHK DDVDAVAKQI 900
901 HQLMENCTRL DVPLLVEVGS GENWDQAH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
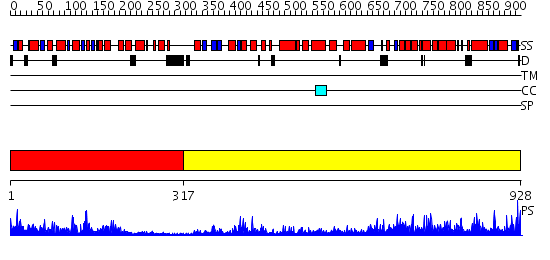
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..316] | 42.30103 | Crystal structure of the human FEN1-PCNA complex | |
| 2 | View Details | [317..928] | 1000.0 | STRUCTURE OF LARGE FRAGMENT OF ESCHERICHIA COLI DNA POLYMERASE I COMPLEXED WITH D/TMP |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.84 |
Source: Reynolds et al. (2008)