













| General Information: |
|
| Name(s) found: |
gi|46581622
[NCBI NR]
gi|46451045 [NCBI NR] |
| Description(s) found:
Found 4 descriptions. SHOW ALL |
|
| Organism: | Desulfovibrio vulgaris str. Hildenborough |
| Length: | 459 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSETTHILVI DDEKNYLLVL EALLTDAGYA VTALNDPETA LAFLEESEVD VVITDMKMPR 60
61 VTGREVLERV KKTWPHIPVC IMTAFGSIEG AVDVMKYGAF DYITKPFSND ELLLSVQNAA 120
121 ELSRVHRQYR ALRANLEERY GTHQIIGRSK AIREVLDMVD RAAPSRSTVL ITGESGTGKE 180
181 LVARAIHFSS PRNSGPFVSV NCMALNPGVL ESELFGHEKG SFTGAMAMRR GRFEQADGGT 240
241 LFLDEIGELT PELQVKLLRV LQERRFERVG GGDEIEVDIR VVAATNKDLQ KEVENGTFRE 300
301 DLYYRLNVVH VALPPLRERR EDIPLLVGHF VEKLARDNGM KPKTFTPEAH DYLTGYEWPG 360
361 NIRQLQNVVE RCLVLVASDT VGVDDLPPEI RDEESQLKSA VDLLPVRLDL ADTLEKIEAA 420
421 LIRRALVRAE FVQVKAAELL GISKSLLQYK LKKYGITGH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
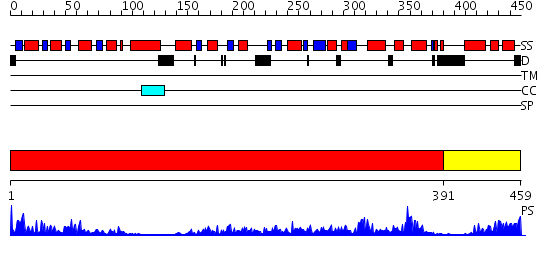
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..390] | 95.0 | Crystal structure of sigm54 activator (AAA+ ATPase) in the inactive state | |
| 2 | View Details | [391..459] | 86.221849 | Crystal structure of a sigma54-activator suggests the mechanism for the conformational switch necessary for sigma54 binding |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)