













| General Information: |
|
| Name(s) found: |
UBP4_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 926 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEQNIISTIR DECIRHRSKY LTIAQLTAIA EAKINEFIIT GKAKDQDLSS LLDKCIDILS 60
61 IYKKNSKDIK NIISCKNKGA MISSNSVMII QLNYVYYKVI HIIVTTNIPH LSEFAKIKLH 120
121 KSTSDEGNGN NNNNEFQLMN IYNTLLETLL KDENIAKIKS FIKSSIKQTK LNHEQEECNL 180
181 MRTGSYITSN QLNSLISSSA NSASSQMEIL LIDIRSRLEF NKSHIDTKNI ICLEPISFKM 240
241 SYSDHDLEKK SLITSPNSEI KMFQSRNLFK FIILYTDANE YNVKQQSVLL DILVNHSFEK 300
301 PISDDFTKIF ILESGFPGWL KSNYGRQVSS SFPSNNNIKD DSVYINGNTS GLSLQHLPKM 360
361 SPSIRHSMDD SMKEMLVAPT PLNHLQQQQQ QQSDNDHVLK RSSSFKKLFS NYTSPNPKNS 420
421 NSNLYSISSL SISSSPSPLP LHSPDPVKGN SLPINYPETP HLWKNSETDF MTNQREQLNH 480
481 NSFAHIAPIN TKAITSPSRT ATPKLQRFPQ TISMNLNMNS NGHSSATSTI QPSCLSLSNN 540
541 DSLDHTDVTP TSSHNYDLDF AVGLENLGNS CYMNCIIQCI LGTHELTQIF LDDSYAKHIN 600
601 INSKLGSKGI LAKYFARLVH MMYKEQVDGS KKISISPIKF KLACGSVNSL FKTASQQDCQ 660
661 EFCQFLLDGL HEDLNQCGSN PPLKELSQEA EARREKLSLR IASSIEWERF LTTDFSVIVD 720
721 LFQGQYASRL KCKVCSHTST TYQPFTVLSI PIPKKNSRNN ITIEDCFREF TKCENLEVDE 780
781 QWLCPHCEKR QPSTKQLTIT RLPRNLIVHL KRFDNLLNKN NDFVIYPFLL DLTPFWANDF 840
841 DGVFPPGVND DELPIRGQIP PFKYELYGVA CHFGTLYGGH YTAYVKKGLK KGWLYFDDTK 900
901 YKPVKNKADA INSNAYVLFY HRVYGV |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
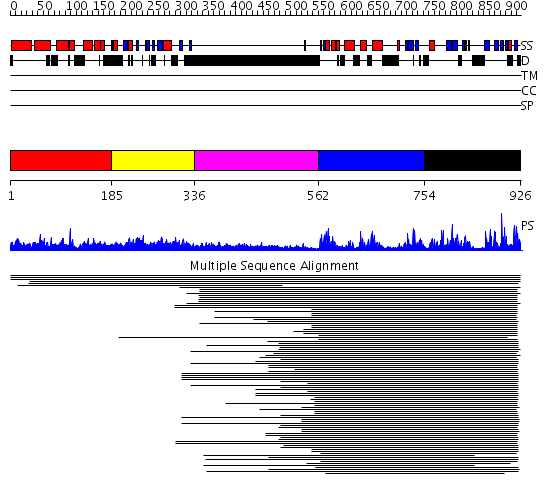
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..184] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [185..335] | 14.0 | Adipocyte-transcription factor FREAC-11 (s12, fkh-14) | |
| 3 | View Details | [336..561] | 7.33 | No description for 1ldvA was found. | |
| 4 | View Details | [562..753] | 3.22999 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [754..926] | 12.214985 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)