













| General Information: |
|
| Name(s) found: |
LCF2_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 744 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAAPDYALTD LIESDPRFES LKTRLAGYTK GSDEYIEELY SQLPLTSYPR YKTFLKKQAV 60
61 AISNPDNEAG FSSIYRSSLS SENLVSCVDK NLRTAYDHFM FSARRWPQRD CLGSRPIDKA 120
121 TGTWEETFRF ESYSTVSKRC HNIGSGILSL VNTKRKRPLE ANDFVVAILS HNNPEWILTD 180
181 LACQAYSLTN TALYETLGPN TSEYILNLTE APILIFAKSN MYHVLKMVPD MKFVNTLVCM 240
241 DELTHDELRM LNESLLPVKC NSLNEKITFF SLEQVEQVGC FNKIPAIPPT PDSLYTISFT 300
301 SGTTGLPKGV EMSHRNIASG IAFAFSTFRI PPDKRNQQLY DMCFLPLAHI FERMVIAYDL 360
361 AIGFGIGFLH KPDPTVLVED LKILKPYAVA LVPRILTRFE AGIKNALDKS TVQRNVANTI 420
421 LDSKSARFTA RGGPDKSIMN FLVYHRVLID KIRDSLGLSN NSFIITGSAP ISKDTLLFLR 480
481 SALDIGIRQG YGLTETFAGV CLSEPFEKDV GSCGAIGISA ECRLKSVPEM GYHADKDLKG 540
541 ELQIRGPQVF ERYFKNPNET SKAVDQDGWF STGDVAFIDG KGRISVIDRV KNFFKLAHGE 600
601 YIAPEKIENI YLSSCPYITQ IFVFGDPLKT FLVGIVGVDV DAAQPILAAK HPEVKTWTKE 660
661 VLVENLNRNK KLRKEFLNKI NKCTDGLQGF EKLHNIKVGL EPLTLEDDVV TPTFKIKRAK 720
721 ASKFFKDTLD QLYAEGSLVK TEKL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
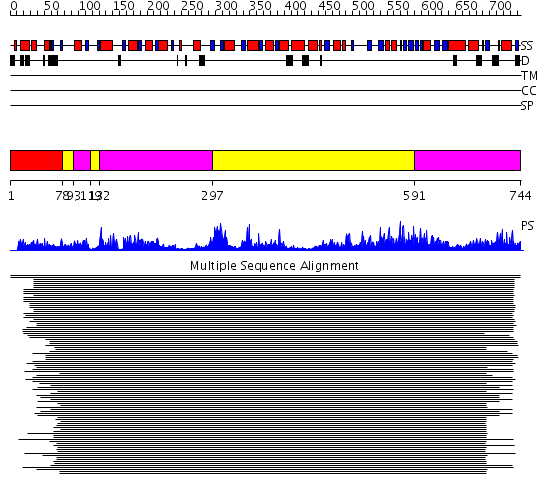
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..77] | N/A | Confident ab initio structure predictions are available. | |
| 2 | View Details | [78..92] [119..131] [297..590] |
348.54902 | Luciferase | |
| 3 | View Details | [93..118] [132..296] [591..744] |
348.54902 | Luciferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)