













| General Information: |
|
| Name(s) found: |
DOT6_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 670 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSISTSLNSA SIHLSSMDTH PQLHSLTRQP HSSSTAMSKN EAQESSPSLP ASSSSSTSAS 60
61 ASASSKNSSK NPSSWDPQDD LLLRHLKEVK KMGWKDISQY FPNRTPNACQ FRWRRLKSGN 120
121 LKSNKTALID INTYTGPLKI THGDETANAQ QKPSKKVEEN VLTEDTAEFT TTSSIPIPSR 180
181 KTSLPSFHAS MSFSQSPSNV TPTTIVSNAA SSMPFAPPTL PAALPHHPHQ HLHHHPHHKT 240
241 LKPRSNSHSF TNSLNQDPIV RSNDEEKYGF IPKVFVRSRR SSFAYPQQVA ITTTPSSPNS 300
301 SHVLLSSKSR RGSLANWSRR SSFNVSSNNT SRRSSMILAP NSVSNIFNVN NSGSNTASTS 360
361 NTNSRRESVI KKEFQQRLNN LSNSGGPTSN NGPIFPNSYT FMDLPHSSSV SSSSTLHKSK 420
421 RGSFSGHSMK SSCNPTNLWS KDEDALLMEN KKRNLSVMEL SILLPQRTEV EIQWRLNALS 480
481 SDADMLSPTH SPQKTLSKKT CPRMFKSGST TDDDKGSDKE DVMGDGSNDD DEDNVDPLHR 540
541 AKQSSNKTVF SSSSSNISSK DVSPDPIFSP DPADDSSNTS DAGSRCTITS DTSSSAATMN 600
601 RTPNSKNPQD IALLNNFRSE AITPRPKPSS TTTSITTETT NNMINHSSST TTTTNNSPLP 660
661 SINTIFKDML |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
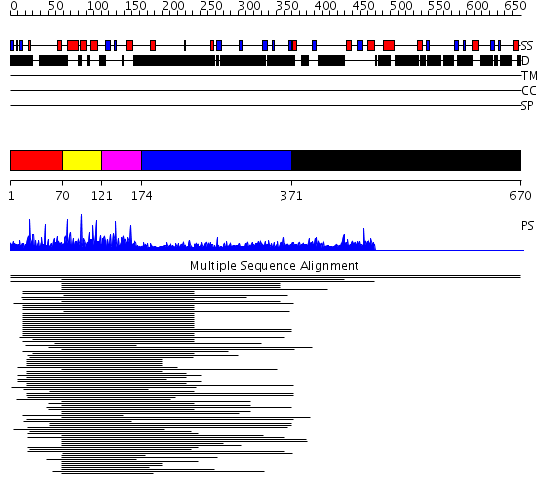
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..69] | 61.221849 | c-Myb, DNA-binding domain | |
| 2 | View Details | [70..120] | 61.221849 | c-Myb, DNA-binding domain | |
| 3 | View Details | [121..173] | 61.221849 | c-Myb, DNA-binding domain | |
| 4 | View Details | [174..370] | 4.301997 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [371..670] | 7.08 | Microbial transglutaminase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)