













| General Information: |
|
| Name(s) found: |
SMC2_YEAST
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1170 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKVEELIIDG FKSYATRTVI TDWDPQFNAI TGLNGSGKSN ILDAICFVLG IASMSTVRAS 60
61 SLQDLIYKRG QAGVTKASVT IVFDNTDKSN SPIGFTNSPQ ISVTRQVVLG GTSKYLINGH 120
121 RAPQQSVLQL FQSVQLNINN PNFLIMQGKI TKVLNMKPSE ILSLIEEAAG TKMFEDRREK 180
181 AERTMSKKET KLQENRTLLT EEIEPKLEKL RNEKRMFLEF QSTQTDLEKT ERIVVSYEYY 240
241 NIKHKHTSIR ETLENGETRM KMLNEFVKKT SEEIDSLNED VEEIKLQKEK ELHKEGTISK 300
301 LENKENGLLN EISRLKTSLS IKVENLNDTT EKSKALESEI ASSSAKLIEK KSAYANTEKD 360
361 YKMVQEQLSK QRDLYKRKEE LVSTLTTGIS STGAADGGYN AQLAKAKTEL NEVSLAIKKS 420
421 SMKMELLKKE LLTIEPKLKE ATKDNELNVK HVKQCQETCD KLRARLVEYG FDPSRIKDLK 480
481 QREDKLKSHY YQTCKNSEYL KRRVTNLEFN YTKPYPNFEA SFVHGVVGQL FQIDNDNIRY 540
541 ATALQTCAGG RLFNVVVQDS QTATQLLERG RLRKRVTIIP LDKIYTRPIS SQVLDLAKKI 600
601 APGKVELAIN LIRFDESITK AMEFIFGNSL ICEDPETAKK ITFHPKIRAR SITLQGDVYD 660
661 PEGTLSGGSR NTSESLLVDI QKYNQIQKQI ETIQADLNHV TEELQTQYAT SQKTKTIQSD 720
721 LNLSLHKLDL AKRNLDANPS SQIIARNEEI LRDIGECENE IKTKQMSLKK CQEEVSTIEK 780
781 DMKEYDSDKG SKLNELKKEL KLLAKELEEQ ESESERKYDL FQNLELETEQ LSSELDSNKT 840
841 LLHNHLKSIE SLKLENSDLE GKIRGVEDDL VTVQTELNEE KKRLMDIDDE LNELETLIKK 900
901 KQDEKKSSEL ELQKLVHDLN KYKSNTNNME KIIEDLRQKH EFLEDFDLVR NIVKQNEGID 960
961 LDTYRERSKQ LNEKFQELRK KVNPNIMNMI ENVEKKEAAL KTMIKTIEKD KMKIQETISK1020
1021 LNEYKRETLV KTWEKVTLDF GNIFADLLPN SFAKLVPCEG KDVTQGLEVK VKLGNIWKES1080
1081 LIELSGGQRS LIALSLIMAL LQFRPAPMYI LDEVDAALDL SHTQNIGHLI KTRFKGSQFI1140
1141 VVSLKEGMFA NANRVFRTRF QDGTSVVSIM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
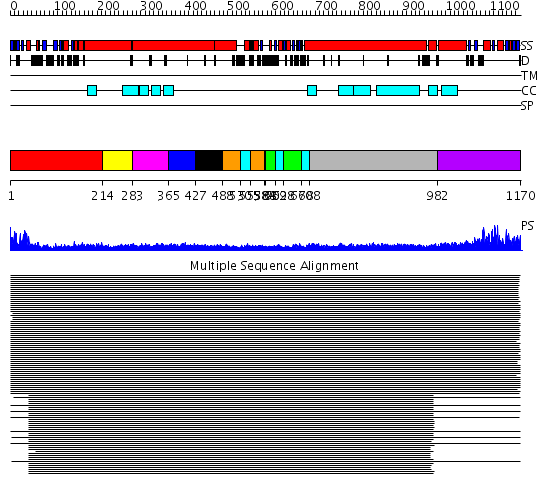
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..213] | 9.69897 | Rad50 | |
| 2 | View Details | [214..282] | 9.69897 | Tropomyosin | |
| 3 | View Details | [283..364] | 9.69897 | Tropomyosin | |
| 4 | View Details | [365..426] | 9.69897 | Tropomyosin | |
| 5 | View Details | [427..487] | 9.69897 | Tropomyosin | |
| 6 | View Details | [488..529] [553..583] |
83.06601 | Smc hinge domain | |
| 7 | View Details | [530..552] [584..587] [609..627] [670..687] |
83.06601 | Smc hinge domain | |
| 8 | View Details | [588..608] [628..669] |
83.06601 | Smc hinge domain | |
| 9 | View Details | [688..981] | 50.9691 | Heavy meromyosin subfragment | |
| 10 | View Details | [982..1170] | 157.68867 | Smc head domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.84 |
Source: Reynolds et al. (2008)