













| General Information: |
|
| Name(s) found: |
SXM1_YEAST
[Swiss-Prot]
|
| Description(s) found:
Found 2 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 944 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVQEQAILSC IEQTMVADAK IIKEAEQQLF EFQKQPGFTS FLLNIVSDDN FALNVRLSSA 60
61 IYLKNKIHRS WDTKREDGIK ADEKLSIKER LIETLVKNCE NNHIRPILTE TINGILVGQE 120
121 DWDLAPIIKN LLSSGDASYI YPGLLLLFQL CKAHRWDMVG SRDYIDSVIE ELFPIVEGIA 180
181 SNIGSQTDYR SNEILYLILK SFKYACLNNL PQYFSQPERI MSWVQLHLYL CSKPLPVEVM 240
241 ELDPADRSLD KRVKVNKWGF GNLNRFLQRY NKITKAITKE FIDYIFNTIV PIILREFFKD 300
301 IEAWGNNSLW LSDSSLYFLI SFLEKCVTID QLYPLIEPHL QIIFENVIFP CLCANEQSIE 360
361 LLEDDQEEYT RRYFDINREG STPDAASADF IFLIGSKRPE KLNNILPFIN DIFTRFDANS 420
421 SDINMAFKEE GALRTLSNLF SFIDEPSVLE NIFGHFIVPL LSQDKYMFLV ARSLETIALY 480
481 SEEFKDMNIL SQLFELTYTN FLNSNVLPVQ IEAADAIKCL IVSNPQIHPA VSAHVPGMME 540
541 KLLKLSKIFE IDILSEVMEA LVERFSDELS PFAKDLASNL VEQFLRIAQA LVENPSETYS 600
601 ASDQEQEIQA SGLLQTMTTM VMSMNKVPLI ESLAPVVKFV VLHAQISFIT EAVDLLDALT 660
661 ISSHLLYNQI APPIWELLHD ILDSFQTYAM DYFEAYSIFF ETIVMTGFPQ DQTYVQPLLE 720
721 ILSAKLESEV DYDIEHVMQI LMYFALSMRD IPLFSKAIKV STNDELGLDS KCIVKLGLAN 780
781 LFAKPIETLQ IMENEGFTIN FFTNWFNEKF YSVFAIKLQV LVILTLLKMP EVPNSVSPLL 840
841 NNLTNKLVEL TLSLPKAIRN RDAVTEGKSL EGDLTPEEEE EYFIECDDDM KETVLDQINV 900
901 FQEVHTFFKN LQNEDAGKYE KIINYLDESK RDSLQVILEF VSQH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
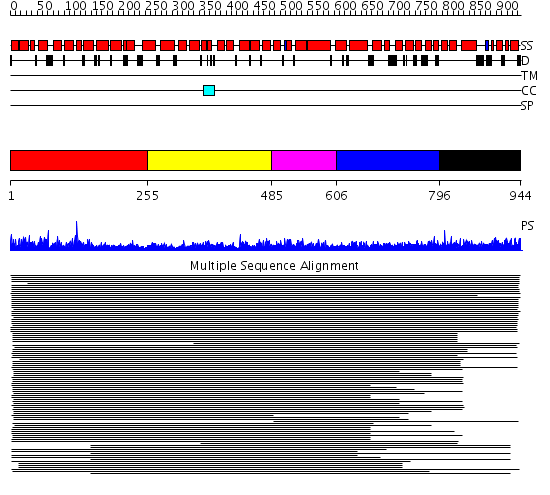
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..254] | 83.69897 | Importin beta | |
| 2 | View Details | [255..484] | 83.69897 | Importin beta | |
| 3 | View Details | [485..605] | 83.69897 | Importin beta | |
| 4 | View Details | [606..795] | 83.69897 | Importin beta | |
| 5 | View Details | [796..944] | 1.053979 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)