













| General Information: |
|
| Name(s) found: |
Ada2b-PB /
FBpp0099776
[FlyBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 555 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
polytene chromosome [IDA] SAGA complex [IPI][NAS] |
| Biological Process: |
positive regulation of histone acetylation
[IMP]
cell proliferation [IDA] histone H3 acetylation [IDA] chromatin remodeling [IDA] eye pigment metabolic process [IDA] regulation of transcription [IDA][IEA] axon target recognition [IMP] |
| Molecular Function: |
histone acetyltransferase activity
[NAS]
DNA binding [IEA] zinc ion binding [IEA] protein binding [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTTIADLFTK YNCTNCQDDI QGIRVHCAEC ENFDLCLQCF AAGAEIGAHQ NNHSYQFMDT 60
61 GTSILSVFRG KGAWTAREEI RLLDAIEQYG FGNWEDISKH IETKSAEDAK EEYVNKFVNG 120
121 TIGRATWTPA QSQRPRLIDH TGDDDAGPLG TNALSTLPPL EINSDEAMQL GYMPNRDSFE 180
181 REYDPTAEQL ISNISLSSED TEVDVMLKLA HVDIYTRRLR ERARRKRMVR DYQLVSNFFR 240
241 NRNYAQQQGL TKEQREFRDR FRVYAQFYTC NEYERLLGSL EREKELRIRQ SELYRYRYNG 300
301 LTKIAECTHF EQHAATATHR STGPYGHGKT DHTHTSNGSH RPPSSSLHSP QPNLRKVEMS 360
361 SGGEASSNSI APRNTLHIAD PTCSGALLPS KNYLDSCRGS SAATMLQTTG MVMGVTVDSG 420
421 ATTGVTSTAT TMANLPTNSA KGSQQHLQPL QQHPQLLQSG NQHKMQNEAA GGGSDQVPSM 480
481 SLKLRTQLEE LKHLPQPPGS ELLSHNELDL CKKHNITPTT YLSVKTVCLS GAPSLGSPME 540
541 TSLRKFFIKC GWLSH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
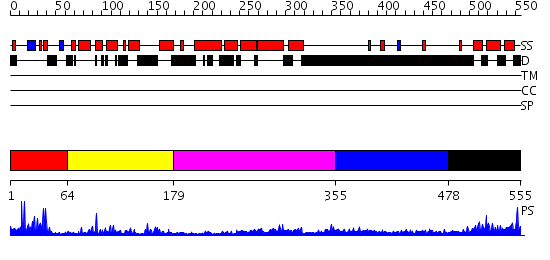
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..63] | 2.87 | No description for 2e5rA was found. | |
| 2 | View Details | [64..178] | 23.221849 | c-Myb, DNA-binding domain | |
| 3 | View Details | [179..354] | 4.470995 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [355..477] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [478..555] | 15.0 | Structural and functional analysis of ada2 alpha swirm domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)