













| General Information: |
|
| Name(s) found: |
CUL4_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 792 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
ubiquitin ligase complex [IPI] CUL4 RING ubiquitin ligase complex [IPI] |
| Biological Process: |
DNA repair
[IMP]
fruit development [IMP] photomorphogenesis [IMP] protein polyubiquitination [IDA] sugar mediated signaling pathway [IMP] cell cycle [ISS] cotyledon development [IMP] hormone-mediated signaling pathway [IMP] flower development [IMP] shoot development [IMP] leaf development [IMP] short-day photoperiodism, flowering [IMP] negative regulation of photomorphogenesis [IMP] |
| Molecular Function: |
protein binding
[IPI]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLPTKRSTF SAASASDDSS YSSPPMKKAK NDLHHSPQHP NTADKVVGFH MEEDPTPAAA 60
61 NLSRKKATLP QPTKKFVIKL NKAKPTLPTN FEENTWEKLQ SAIRAIFLKK KISFDLESLY 120
121 QAVDNLCLHK LDGKLYDQIE KECEEHISAA LQSLVGQNTD LTVFLSRVEK CWQDFCDQML 180
181 MIRSIALTLD RKYVIQNPNV RSLWEMGLQL FRKHLSLAPE VEQRTVKGLL SMIEKERLAE 240
241 AVNRTLLSHL LKMFTALGIY MESFEKPFLE GTSEFYAAEG MKYMQQSDVP EYLKHVEGRL 300
301 HEENERCILY IDAVTRKPLI TTVERQLLER HILVVLEKGF TTLMDGRRTE DLQRMQTLFS 360
361 RVNALESLRQ ALSSYVRKTG QKIVMDEEKD KDMVQSLLDF KASLDIIWEE SFYKNESFGN 420
421 TIKDSFEHLI NLRQNRPAEL IAKFLDEKLR AGNKGTSEEE LESVLEKVLV LFRFIQGKDV 480
481 FEAFYKKDLA KRLLLGKSAS IDAEKSMISK LKTECGSQFT NKLEGMFKDI ELSKEINESF 540
541 KQSSQARTKL PSGIEMSVHV LTTGYWPTYP PMDVKLPHEL NVYQDIFKEF YLSKYSGRRL 600
601 MWQNSLGHCV LKADFSKGKK ELAVSLFQAV VLMLFNDAMK LSFEDIKDST SIEDKELRRT 660
661 LQSLACGKVR VLQKNPKGRD VEDGDEFEFN DEFAAPLYRI KVNAIQMKET VEENTSTTER 720
721 VFQDRQYQID AAIVRIMKTR KVLSHTLLIT ELFQQLKFPI KPADLKKRIE SLIDREYLER 780
781 EKSNPQIYNY LA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
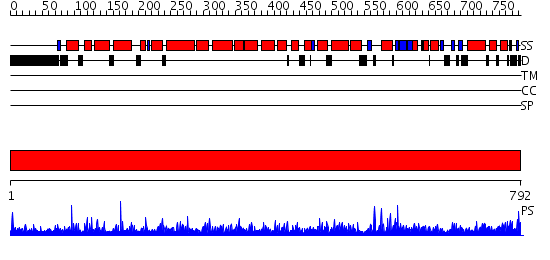
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..792] | 1000.0 | No description for 2hyeC was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.57 |
Source: Reynolds et al. (2008)