













| General Information: |
|
| Name(s) found: |
WNK8_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 563 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
|
| Biological Process: |
photoperiodism, flowering
[IMP]
protein amino acid autophosphorylation [IDA] vegetative to reproductive phase transition of meristem [IMP] protein amino acid phosphorylation [IDA][TAS] |
| Molecular Function: |
protein kinase activity
[IDA][TAS]
kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASGSGFLGQ ISSMEEADFA EKDPSGRYIR YDDVLGRGAF KTVYKAFDEV DGIEVAWNLV 60
61 SIEDVMQMPG QLERLYSEVH LLKALKHENI IKLFYSWVDE KNKTINMITE LFTSGSLRVY 120
121 RKKHRKVDPK AIKNWARQIL KGLNYLHSQN PPVIHRDLKC DNIFVNGNTG EVKIGDLGLA 180
181 TVLQQPTARS VIGTPEFMAP ELYEEEYNEL VDIYSFGMCM LEMVTCEYPY NECRNQAQIY 240
241 KKVTSNIKPQ SLGKVDDPQV RQFIEKCLLP ASSRPTALEL SKDPFLARDG GKDSALLASS 300
301 STSSKYVRPP QLEHLPMDVD HNENKSVSSN EDYPWSQTIE LQRIAENKEF RLRGERSDDV 360
361 TASMVLRIAD PSGKCRIVHF AFYLESDTAT AIAEEMVEEL HLTSQEVVVI ADMIDDFIMQ 420
421 LLSDRTSSHH NQNSPRLTHE DHEAANQQTV NSKDEEAAGQ SMKSDISADY YFPYSANDGN 480
481 AAMEAGRDAE SMSSYLDSCS MMSTIYNLSI SDNDYPEDLK TELNLIESQF NQSFQDLLKL 540
541 KEDAIENAKR KWITKKQKAV NIS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
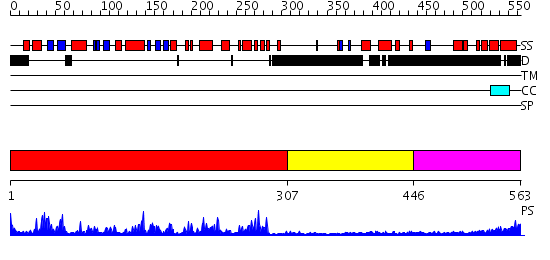
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..306] | 59.522879 | Crystal structure of the kinase domain of WNK1, a kinase that causes a hereditary form of hypertension | |
| 2 | View Details | [307..445] | 2.21 | No description for 2v3sA was found. | |
| 3 | View Details | [446..563] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)