













| General Information: |
|
| Name(s) found: |
RQL3_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 713 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
DNA recombination
[IEA]
|
| Molecular Function: |
ATP binding
[ISS]
ATP-dependent helicase activity [IDA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKKSPLPVQN VQSSDKNVAG KEALVKLLRW HFGHADFRGK QLEAIQAVVS GRDCFCLMPT 60
61 GGGKSICYQI PALAKPGIVL VVSPLIALME NQVMALKEKG IAAEYLSSTQ ATHVKNKIHE 120
121 DLDSGKPSVR LLYVTPELIA TKGFMLKLRK LHSRGLLNLI AIDEAHCISS WGHDFRPSYR 180
181 QLSTLRDSLA DVPVLALTAT AAPKVQKDVI DSLNLRNPLV LKSSFNRPNI FYEVRYKDLL 240
241 DNAYTDLGNL LKSCGNICAI IYCLERTTCD DLSVHLSSIG ISSAAYHAGL NSKMRSTVLD 300
301 DWLSSKKQII VATVAFGMGI DKKDVRMVCH FNIPKSMESF YQESGRAGRD QLPSRSVLYY 360
361 GVDDRKKMEY LLRNSENKKS SSSKKPTSDF EQIVTYCEGS GCRRKKILES FGEEFPVQQC 420
421 KKTCDACKHP NQVAHCLEEL MTTASRRHNS SRIFITSSNN KTNEGQYSEF WNRNEDGSNS 480
481 NEEISDSDDA TEAANSVTGP KLSKKLGLDE KLVLLEQAEE KYYERNKQVK KSEKNAISEA 540
541 LRDSSKQRLL DALTRVLQLL ACVEEIDSQK GSEFLENECY RKYSKAGKSF YYSQIASTVR 600
601 WLGTASRDEL MTRLSSVVSL AREQEPLEEP LLATEPVENI EEEDDGKTNT VESRVDEPTQ 660
661 ELVVSPILSP IRLPQVPSFS EFVNRRKIKQ NTLIDKSSEG FDDKKPAKIM KLQ |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
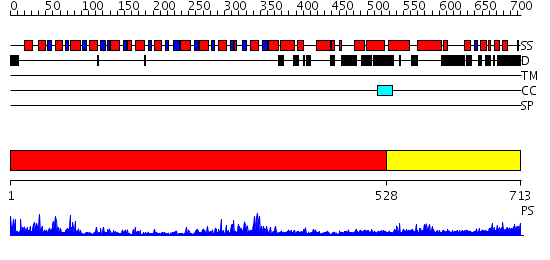
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..527] | 100.0 | Structure of the RecQ Catalytic Core | |
| 2 | View Details | [528..713] | 12.09691 | Crystal structure of SecA:ADP in an open conformation from Bacillus Subtilis |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)