













| General Information: |
|
| Name(s) found: |
gi|221312970
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Bacillus subtilis subsp. subtilis str. NCIB 3610 |
| Length: | 605 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNSVPNDLQT WKRFVKDGVL DEARLRKRIA ESWHRCKKAE VNPYLEKGPK VLQQTELDQQ 60
61 SKKHSFFLTT AKPYLEKLLP AIKEMEMMAL LIDSDGVVLA LDGHPRALYE AKRINFVEGA 120
121 CWTETAVGTN AIGTALHISE PVAIQGSEHY SIASHLWNCS AAPIHHEDGS LAGVIDISCP 180
181 AAGAHPHMLG IATAIAYAAE RELAAKSREK ELELISRFGE RAASSVPMVL CNTKQHIISA 240
241 SMPIRTSMPD WQGRHLYELK ERGYSIENAV TIGDGGTCFY LSEQKKKKAF RFNGVIGQSG 300
301 RSQAMLMHLE RAAATDASVC LSGETGTGKE VAARALHENS ERRHGPFVAV NCGAIPSDLI 360
361 ESELFGYAEG AFTGAKRNGY KGAFQKANQG TLFLDEIGEI SHSMQVALLR VLQERKITPI 420
421 GGTKEIPVDI RVIAATHCDL RELAENGKIR EDLFYRLHVY PIELPPLRDR TEDIPDLFEY 480
481 YKQKNHWPGD LPSDFCNVLK QWKWPGNIRE LFNVFERLSI RFPDGRLRDE SLPALLEAAG 540
541 LPASSAEKKP AAAGVLTFRE QIQKDMMIKA LESAKGNVSQ AAKISGIPRS TFYKRLKKFN 600
601 LSAES |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
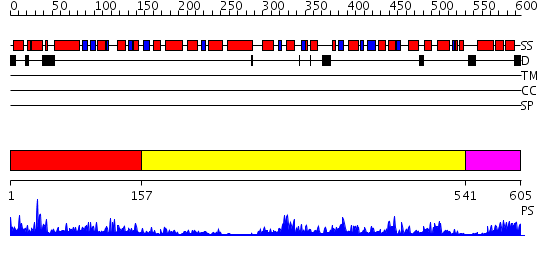
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..156] | 4.948982 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [157..540] | 82.39794 | Crystal structure of sigm54 activator (AAA+ ATPase) in the inactive state | |
| 3 | View Details | [541..605] | 80.39794 | Crystal structure of a sigma54-activator suggests the mechanism for the conformational switch necessary for sigma54 binding |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)