













| General Information: |
|
| Name(s) found: |
TGL5 /
YOR081C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 749 amino acids |
Gene Ontology: |
|
| Cellular Component: |
lipid particle
[IDA |
| Biological Process: |
triglyceride mobilization
[IDA |
| Molecular Function: |
triglyceride lipase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNTLPVTEF LLSKYYELSN TPATDSSSLF KWLYHKTLSR KQLLISDLSS QKKHAISYDQ 60
61 WNDIASRLDD LTGLSEWKTI DESSLYNYKL LQDLTIRMRH LRTTHDYHRL LYLIRTKWVR 120
121 NLGNMNNVNL YRHSHTGTKQ IIHDYLEESQ AVLTALIHQS NMNDHYLLGI LQQTRRNIGR 180
181 TALVLSGGST FGLFHIGVLA ALFESDLMPK VISGSSAGAI VASIFCVHTT QEIPSLLTNV 240
241 LNMEFNIFND DNSKSPNENL LIKISRFCQN GTWFNNQPLI NTMLSFLGNL TFREAYNKTG 300
301 KILNITVSPA SIYEQPKLLN NLTAPNVLIW SAVCASCSLP GVFPSTPLFE KDPHTGKIKE 360
361 WGATNLHLSN MKFMDGSVDN DMPISRLSEM FNVDHIIACQ VNIHVFPLLK FSNTCVGGEI 420
421 EKEITARFRN QVTKIFKFFS DETIHFLDIL KELEFHPYLM TKLKHLFLQQ YSGNVTILPD 480
481 LSMVGQFHEV LKNPSQLFLL HQTTLGARAT WPKISMIQNN CGQEFALDKA ITFLKEKIII 540
541 SSSIKNPLQF YQPRFSEQIK SLSIMDADLP GVDLEESSSN SLSIIKSPNK TAAPGRFPLQ 600
601 PLPSPSSTFN KRKMDMLSPS PSPSTSPQRS KSSFTQQGTR QKANSLSFAI GASSLRLKKS 660
661 PLKVPSRPQF KKRSSYYNQN MSAEMRKNRK KSGTISSYDV QTNSEDFPIP AIENGSFDNT 720
721 LFNPSRFPMD AMSAATNDNF MNNSDIFQN |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
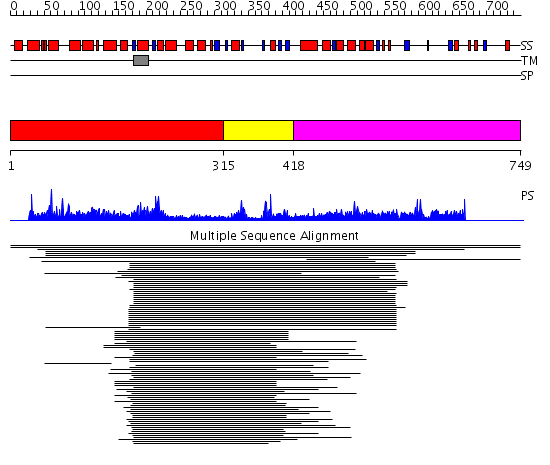
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..314] | 7.19 | Monomeric isocitrate dehydrogenase | |
| 2 | View Details | [315..417] | 2.157991 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [418..749] | 1.075883 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [572..749] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|