













| General Information: |
|
| Name(s) found: |
MET5 /
YJR137C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1442 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA sulfite reductase complex (NADPH) [IDA |
| Biological Process: |
response to drug
[IMP cellular cell wall organization [IMP cellular amino acid biosynthetic process [TAS sulfate assimilation [NAS |
| Molecular Function: |
sulfite reductase (NADPH) activity
[NAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTASDLLTLP QLLAQYSSSA PQNKVFYTTS TKNSHSSFKG LESVATDATH LLNNQDPLNT 60
61 IKDQLSKDIL TTVFTDETTL VKSIHHLYSL PNKLPLVITV DLNLQDYSAI PALKDLSFPI 120
121 LISSDLQTAI SNADSSYKIA TSSLTPVFHF LNLEKIGTST AIEQDIDFPT LEIANEETKV 180
181 ALSEATDSLT NFELVKGKES ITTVIVNLSP YDAEFSSVLP SNVGLIKIRV YRPWNFSKFL 240
241 EILPSSVTKI AVLQGVSKKS QSNEFQPFLL DFFGNFNELV SRNIEQVVLT NIGNVNDYGN 300
301 VINTVISNIN KKEPDNNLFL GESNEKAEEQ AEVTQLISSV KKVVNLEDAY IKVLKQLFSS 360
361 NLQILNQFSS ETIEPSNPEF GFGRFLKQEA QREELISLAK TSLDPSLYLS EDANKIVQLL 420
421 SKWLSFNGRD LDEAQLQEAN ATGLEIFQLL QSNQDSSTVL KFLKIAPTSD SFIFKSSWLI 480
481 GSDAWSYDLG HSGIQQVLSS RKNINVLLID SEPYDHRKQN QDRKKDVGLY AMNYYSAYVA 540
541 SVAVYASYTQ LLTAIIEASK YNGPSIVLAY LPYNSENDTP LEVLKETKNA VESGYWPLYR 600
601 FNPVYDDPST DKEAFSLDSS VIRKQLQDFL DRENKLTLLT RKDPSLSRNL KQSAGDALTR 660
661 KQEKRSKAAF DQLLEGLSGP PLHVYYASDG GNAANLAKRL AARASARGLK ATVLSMDDII 720
721 LEELPGEENV VFITSTAGQG EFPQDGKSFW EALKNDTDLD LASLNVAVFG LGDSEYWPRK 780
781 EDKHYFNKPS QDLFKRLELL SAKALIPLGL GDDQDADGFQ TAYSEWEPKL WEALGVSGAA 840
841 VDDEPKPVTN EDIKRESNFL RGTISENLKD TSSGGVTHAN EQLMKFHGIY TQDDRDIREI 900
901 RKSQGLEPYY MFMARARLPG GKTTPQQWLA LDHLSDTSGN GTLKLTTRAT FQIHGVLKKN 960
961 LKHTLRGMNA VLMDTLAAAG DVNRNVMVSA LPTNAKVHQQ IADMGKLISD HFLPKTTAYH1020
1021 EVWLEGPEEQ DDDPSWPSIF ENRKDGPRKK KTLVSGNALV DIEPIYGPTY LPRKFKFNIA1080
1081 VPPYNDVDVL SIDVGLVAIV NPETQIVEGY NVFVGGGMGT THNNKKTYPR LGSCLGFVKT1140
1141 EDIIPPLEGI VIVQRDHGDR KDRKHARLKY TVDDMGVEGF KQKVEEYWGK KFEPERPFEF1200
1201 KSNIDYFGWI KDETGLNHFT AFIENGRVED TPDLPQKTGI RKVAEYMLKT NSGHFRLTGN1260
1261 QHLVISNITD EHVAGIKSIL KTYKLDNTDF SGLRLSSSSC VGLPTCGLAF AESERFLPDI1320
1321 ITQLEDCLEE YGLRHDSIIM RMTGCPNGCS RPWLGELALV GKAPHTYNLM LGGGYLGQRL1380
1381 NKLYKANVKD EEIVDYIKPL FKRYALEREE GEHFGDFCIR VGIIKPTTEG KYFHEDVSED1440
1441 AY |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |

|
MET10 MET5 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | 3829 - low filter | Green EM, et al (2005) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
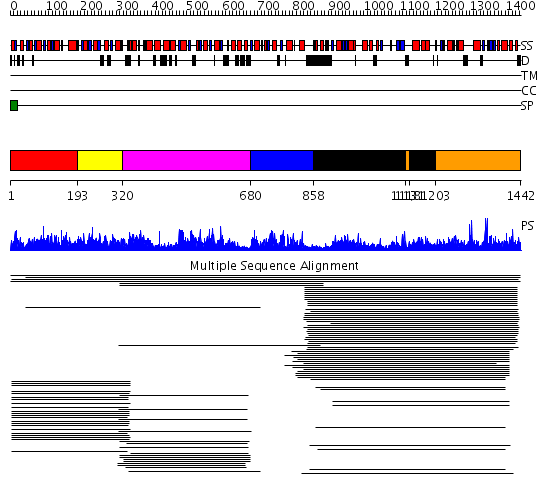
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..192] | 13.18 | E1-beta subunit of pyruvate dehydrogenase, Pyr module; E1-beta subunit of pyruvate dehydrogenase, C-domain | |
| 2 | View Details | [193..319] | 13.18 | E1-beta subunit of pyruvate dehydrogenase, Pyr module; E1-beta subunit of pyruvate dehydrogenase, C-domain | |
| 3 | View Details | [320..679] | 411.228787 | Pyruvate-ferredoxin oxidoreductase, PFOR, domain I; Pyruvate-ferredoxin oxidoreductase, PFOR, domains VI; Pyruvate-ferredoxin oxidoreductase, PFOR, domain II; Pyruvate-ferredoxin oxidoreductase, PFOR, domain III; Pyruvate-ferredoxin oxidoreductase, PFOR, domain V | |
| 4 | View Details | [680..857] | 191.9691 | NADPH-cytochrome p450 reductase, N-terminal domain | |
| 5 | View Details | [858..1117] [1131..1202] |
1386.0 | Sulfite reductase, domains 1 and 3; Sulfite reductase hemoprotein (SiRHP), domains 2 and 4 | |
| 6 | View Details | [1118..1130] [1203..1442] |
1386.0 | Sulfite reductase, domains 1 and 3; Sulfite reductase hemoprotein (SiRHP), domains 2 and 4 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||
| 4 |
|
||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 6 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|