













| General Information: |
|
| Name(s) found: |
UBA4 /
YHR111W
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 440 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
response to oxidative stress
[IMP protein modification process [IDA pseudohyphal growth [IMP cell budding [IGI invasive growth in response to glucose limitation [IGI |
| Molecular Function: |
URM1 activating enzyme activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNDYHLEDTT SELEALRLEN AQLREQLAKR EDSSRDYPLS LEEYQRYGRQ MIVEETGGVA 60
61 GQVKLKNTKV LVVGAGGLGC PALPYLAGAG VGQIGIVDND VVETSNLHRQ VLHDSSRVGM 120
121 LKCESARQYI TKLNPHINVV TYPVRLNSSN AFDIFKGYNY ILDCTDSPLT RYLVSDVAVN 180
181 LGITVVSASG LGTEGQLTIL NFNNIGPCYR CFYPTPPPPN AVTSCQEGGV IGPCIGLVGT 240
241 MMAVETLKLI LGIYTNENFS PFLMLYSGFP QQSLRTFKMR GRQEKCLCCG KNRTITKEAI 300
301 EKGEINYELF CGARNYNVCE PDERISVDAF QRIYKDDEFL AKHIFLDVRP SHHYEISHFP 360
361 EAVNIPIKNL RDMNGDLKKL QEKLPSVEKD SNIVILCRYG NDSQLATRLL KDKFGFSNVR 420
421 DVRGGYFKYI DDIDQTIPKY |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
GUS1 QRI1 SLX4 UBA4 YOR097C |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
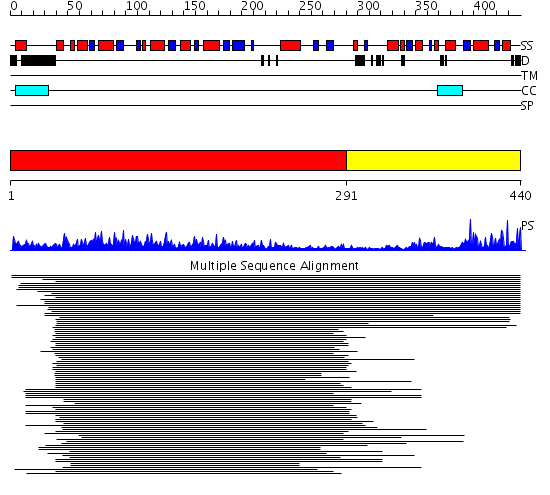
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..290] | 443.9691 | Molybdenum cofactor biosynthesis protein MoeB | |
| 2 | View Details | [291..440] | 17.154902 | Sulfurtransferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.83 |
Source: Reynolds et al. (2008)