













| General Information: |
|
| Name(s) found: |
RRM3 /
YHR031C
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 723 amino acids |
Gene Ontology: |
|
| Cellular Component: |
replication fork
[IDA nuclear telomeric heterochromatin [IPI |
| Biological Process: |
DNA replication
[IGI mitochondrial genome maintenance [IGI |
| Molecular Function: |
DNA helicase activity
[IMP ATP-dependent DNA helicase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFRSHASGNK KQWSKRSSNG STPAASASGS HAYRQQTLSS FFMGCGKKSA AASKNSTTII 60
61 DLESGDEGNR NITAPPRPRL IRNNSSSLFS QSQGSFGDDD PDAEFKKLVD VPRLNSYKKS 120
121 SRSLSMTSSL HKTASASTTQ KTYHFDEDET LREVTSVKSN SRQLSFTSTI NIEDSSMKLS 180
181 TDSERPAKRS KPSMEFQGLK LTVPKKIKPL LRKTVSNMDS MNHRSASSPV VLTMEQERVV 240
241 NLIVKKRTNV FYTGSAGTGK SVILQTIIRQ LSSLYGKESI AITASTGLAA VTIGGSTLHK 300
301 WSGIGIGNKT IDQLVKKIQS QKDLLAAWRY TKVLIIDEIS MVDGNLLDKL EQIARRIRKN 360
361 DDPFGGIQLV LTGDFFQLPP VAKKDEHNVV KFCFESEMWK RCIQKTILLT KVFRQQDNKL 420
421 IDILNAIRYG ELTVDIAKTI RNLNRDIDYA DGIAPTELYA TRREVELSNV KKLQSLPGDL 480
481 YEFKAVDNAP ERYQAILDSS LMVEKVVALK EDAQVMMLKN KPDVELVNGS LGKVLFFVTE 540
541 SLVVKMKEIY KIVDDEVVMD MRLVSRVIGN PLLKESKEFR QDLNARPLAR LERLKILINY 600
601 AVKISPHKEK FPYVRWTVGK NKYIHELMVP ERFPIDIPRE NVGLERTQIP LMLCWALSIH 660
661 KAQGQTIQRL KVDLRRIFEA GQVYVALSRA VTMDTLQVLN FDPGKIRTNE RVKDFYKRLE 720
721 TLK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
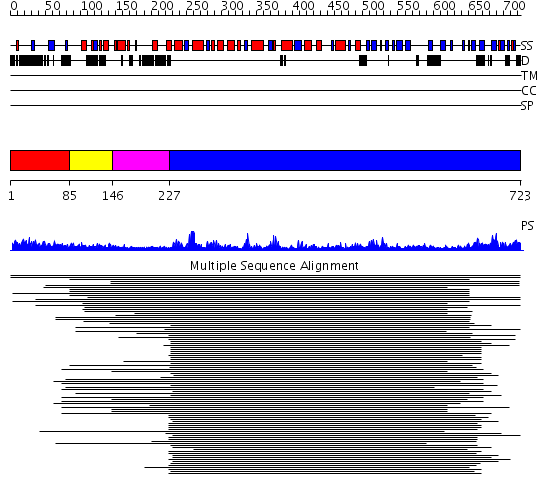
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..84] | 2.129989 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [85..145] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [146..226] | 21.154902 | Signal recognition particle receptor, FtsY; GTPase domain of the signal recognition particle receptor FtsY | |
| 4 | View Details | [227..723] | 63.045757 | DEXX box DNA helicase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|