













| General Information: |
|
| Name(s) found: |
raf1 /
SPCC613.12c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 638 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mating-type region heterochromatin
[TAS nucleus [IDA CLRC ubiquitin ligase complex [IDA nuclear centromeric heterochromatin [IC cytosol [IDA |
| Biological Process: |
meiotic telomere clustering
[IMP positive regulation of histone H3-K9 methylation [IMP chromatin silencing at centromere [IMP negative regulation of histone H3-K4 methylation [IMP regulation of mating type switching [IMP meiotic chromosome segregation [IMP mitotic sister chromatid segregation [IMP chromatin silencing at silent mating-type cassette [IMP chromatin silencing at telomere [IMP meiotic sister chromatid cohesion, centromeric [IMP cellular protein localization [IGI chromatin silencing by small RNA [IMP |
| Molecular Function: |
protein binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTNSSPRVKR RTDTQYLHSV SSKLPKVDFD TNNEEFFEEE FEIYDPFYRA ELPCPKPSLS 60
61 ISKHSIAKVP SNVNKRLELQ LLLTSGTFLP NSRPYLSERV RKHTHLLSNS ITGDDKPSLI 120
121 HVDFTPEECF ILQEAKLKFG PVNSVQFNDA YSTHISPKLP GRAYEDCQKF EIDNPSLSPV 180
181 DKHGAIILRT YKKNKKLLPD YLKSFYNAGS SYFQREQVHQ LMDGESVFFL KPWKHFNETS 240
241 GDTVCVAYNP LCEKFALGST AQDGAYNRLG NLWIGDFHSE TIQSLESHYK LNQVGEKEYS 300
301 TISDLCFSKG NLFLYTGAFD NAVKVWDMEG NLCGIFNAPT DYIHKLALSD DDLLAVACKN 360
361 GYGYLLSTDN STGEILTSAN LIYPEALEKG YSASLIEFSN FLGRSSDKVI IGYDSFHTSN 420
421 NRGCLALFDA STASFVQKFN TADEAFTSLY MHPSQVGFVA SSNTLSNGRV YYLDTRMYKV 480
481 CLNFTTTQKD INHATISNSG ILVTSSGTDN QTFVWDSRKP DKPLSLLKHG KTKMIHFDGA 540
541 NEEEVDAGIN MAQWQPKGNL FVTGGSDGIV KVWDLRLNNP FIQNFTEMNS AITYGGFSED 600
601 ASKLTVCCVG GDVNMYSLGN DNGNKFGEFR IIENRLLT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
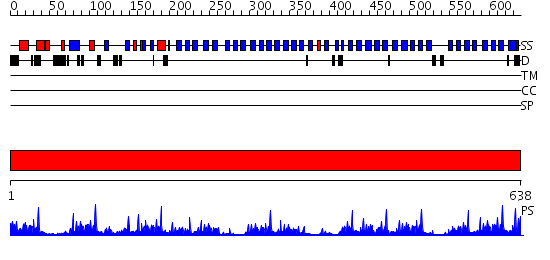
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..638] | 32.221849 | Actin interacting protein 1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)