













| General Information: |
|
| Name(s) found: |
FBpp0309142
[FlyBase]
FBpp0309141 [FlyBase] FBpp0309140 [FlyBase] r-PA / FBpp0088675 [FlyBase] |
| Description(s) found:
Found 33 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 2224 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[NAS]
|
| Biological Process: |
'de novo' pyrimidine base biosynthetic process
[IMP][NAS]
glutamine metabolic process [IC][IMP] |
| Molecular Function: |
ATP binding
[IC][IEA]
aspartate carbamoyltransferase activity [IDA][IMP][ISS] dihydroorotase activity [IDA][IMP][ISS] carbamoyl-phosphate synthase (ammonia) activity [ISS] carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity [IDA][IMP] amino acid binding [IEA] glutamine binding [IC] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASTDCYLAL EDGTVLPGYS FGYVPSENES KVGFGGEVVF QTGMVGYTEA LTDRSYSGQI 60
61 LVLTYPLIGN YGVPAPDEDE HGLPLHFEWM KGVVQATALV VGEVAEEAFH WRKWKTLPDW 120
121 LKQHKVPGIQ DIDTRALTKK LREQGSMLGK IVYEKPPVEG LPKSSFVDPN VRNLAKECSV 180
181 KERQVYGNPN GKGPRIAILD CGLKLNQLRC LLQRGASVTL LPWSARLEDE QFDALFLSNG 240
241 PGNPESCDQI VQQVRKVIEE GQKPVFGICL GHQLLAKAIG CSTYKMKYGN RGHNLPCLHR 300
301 ATGRCLMTSQ NHGYAVDLEQ LPDGWSELFV NANDGTNEGI VHASKPYFSV QFHPEHHAGP 360
361 QDTEFLFDVF MESIQQKDLT IPQLIEQRLR PTTPAIDSAP VMPRKVLILG SGGLSIGQAG 420
421 EFDYSGSQAI KAMRESNIQT VLINPNIATV QTSKGMADKC YFLPLTPHYV EQVIKSERPN 480
481 GVLLTFGGQT ALNCGVQLER AGVFSKYNVR ILGTPIQSII ETEDRKLFAE RVNEIGEQVA 540
541 PSEAVYSVAQ ALDAASRLGY PVMARAAFSL GGLGSGFANN EEELQSLAQQ ALAHSSQLIV 600
601 DKSLKGWKEV EYEVVRDAYN NCITVCNMEN FDPLGIHTGE SIVVAPSQTL SDREYQMLRS 660
661 TALKVIRHFG VVGECNIQYA LCPHSEQYYI IEVNARLSRS SALASKATGY PLAYVAAKLA 720
721 LGLPLPDIKN SVTGNTTACF EPSLDYCVVK IPRWDLAKFV RVSKHIGSSM KSVGEVMAIG 780
781 RNFEEAFQKA LRMVDSDVLG FDPDVVPLNK EQLAEQLSEP TDRRPFVIAA ALQLGMSLRE 840
841 LHQLTNIDYW FLEKLERIIL LQSLLTRNGS RTDAALLLKA KRFGFSDKQI AKYIKSTELA 900
901 VRHQRQEFGI RPHVKQIDTV AGEWPASTNY LYHTYNGSEH DVDFPGGHTI VVGSGVYRIG 960
961 SSVEFDWCAV GCLRELRKLQ RPTIMINYNP ETVSTDYDMC DRLYFEEISF EVVMDIYEME1020
1021 NSEGIILSMG GQLPNNIAMD LHRQQAKVLG TSPESIDCAE NRFKFSRMLD RKGILQPRWK1080
1081 ELTNLQSAIE FCEEVGYPCL VRPSYVLSGA AMNVAYSNQD LETYLNAASE VSREHPVVIS1140
1141 KFLTEAKEID VDAVASDGRI LCMAVSEHVE NAGVHSGDAT LVTPPQDLNA ETLEAIKRIT1200
1201 CDLASVLDVT GPFNMQLIAK NNELKVIECN VRVSRSFPFV SKTLDHDFVA TATRAIVGLD1260
1261 VEPLDVLHGV GKVGVKVPQF SFSRLAGADV QLGVEMASTG EVACFGDNRY EAYLKAMMST1320
1321 GFQIPKNAVL LSIGSFKHKM ELLPSIRDLA KMGYKLYASM GTGDFYAEHG VNVESVQWTF1380
1381 DKTTPDDING ELRHLAEFLA NKQFDLVINL PMSGGGARRV SSFMTHGYRT RRLAVDYSIP1440
1441 LVTDVKCTKL LVESMRMNGG KPPMKTHTDC MTSRRIVKLP GFIDVHVHLR EPGATHKEDF1500
1501 ASGTAAALAG GVTLVCAMPN TNPSIVDRET FTQFQELAKA GARCDYALYV GASDDNWAQV1560
1561 NELASHACGL KMYLNDTFGT LKLSDMTSWQ RHLSHWPKRS PIVCHAERQS TAAVIMLAHL1620
1621 LDRSVHICHV ARKEEIQLIR SAKEKGVKVT CEVCPHHLFL STKDVERLGH GMSEVRPLLC1680
1681 SPEDQEALWE NIDYIDVFAT DHAPHTLAEK RSERPPPGFP GVETILPLLL QAVHEGRLTM1740
1741 EDIKRKFHRN PKIIFNLPDQ AQTYVEVDLD EEWTITGNEM KSKSGWTPFE GTKVKGRVHR1800
1801 VVLRGEVAFV DGQVLVQPGF GQNVRPKQSP LASEASQDLL PSDNDANDTF TRLLTSEGPG1860
1861 GGVHGISTKV HFVDGANFLR PNSPSPRIRL DSASNTTLRE YLQRTTNSNP VAHSLMGKHI1920
1921 LAVDMFNKDH LNDIFNLAQL LKLRGTKDRP VDELLPGKIM ASVFYEVSTR TQCSFAAAML1980
1981 RLGGRVISMD NITSSVKKGE SLEDSIKVVS SYADVVVLRH PSPGAVARAA TFSRKPLINA2040
2041 GDGVGEHPTQ ALLDIFTIRE EFGTVNGLTI TMVGDLKNGR TVHSLARLLT LYNVNLQYVA2100
2101 PNSLQMPDEV VQFVHQRGVK QLFARDLKNV LPDTDVLYMT RIQRERFDNV EDYEKCCGHL2160
2161 VLTPEHMMRA KKRSIVLHPL PRLNEISREI DSDPRAAYFR QAEYGMYIRM ALLAMVVGGR2220
2221 NTAL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
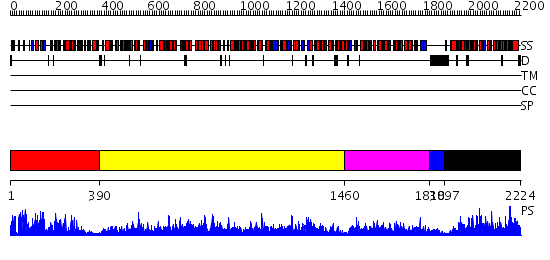
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..389] | 125.0 | Carbamoyl phosphate synthetase, small subunit N-terminal domain; Carbamoyl phosphate synthetase, small subunit C-terminal domain | |
| 2 | View Details | [390..1459] | 1000.0 | Carbamoyl phosphate synthetase, large subunit connection domain; Carbamoyl phosphate synthetase, large subunit allosteric, C-terminal domain; Carbamoyl phosphate synthetase (CPS), large subunit PreATP-grasp domains; Carbamoyl phosphate synthetase (CPS), large subunit ATP-binding domains | |
| 3 | View Details | [1460..1829] | 63.69897 | The Crystal Structure of a Novel, Latent Dihydroorotase from Aquifex aeolicus at 1.7 A resolution | |
| 4 | View Details | [1830..1896] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [1897..2224] | 98.221849 | Aspartate carbamoyltransferase catalytic subunit |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)