













| General Information: |
|
| Name(s) found: |
CG11198-PA /
FBpp0087947
[FlyBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 2482 amino acids |
Gene Ontology: |
|
| Cellular Component: |
lipid particle
[IDA]
|
| Biological Process: |
fatty acid biosynthetic process
[IEA]
|
| Molecular Function: |
ATP binding
[IEA]
acetyl-CoA carboxylase activity [ISS] biotin binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLITILGTLA AFLAFLLTLI FGRGQKRSKP VQSSAATSAT TATTGDSDNG NTNHNSSVIA 60
61 ATATATTTSS KPPIAPAAPP SAVKASDKRF PKACIKKVQF SSESLRSDVD ELCDQLKDSA 120
121 LSNLSNDNIR IACHQNNNNS SINKNQNNNS IDISISISKM SETNESNDTA AQSAEGERPS 180
181 FLVGDEIDER AAEAGEACDE FPLKMQNDVR QNGDISERRK RLRPSMSRGT GLGQDRHQDR 240
241 DFHIATTEEF VKRFGGTRVI NKVLIANNGI AAVKCMRSIR RWAYEMFKNE RAIRFVVMVT 300
301 PEDLKANAEY IKMADHYVPV PGGSNNNNYA NVELIVDIAL RTQVQAVWAG WGHASENPKL 360
361 PELLHKEGLV FLGPPERAMW ALGDKVASSI VAQTAEIPTL PWSGSDLKAQ YSGKKIKISS 420
421 ELFARGCVTN VEQGLAAVNK IGFPVMIKAS EGGGGKGIRR VDTTEEFPGL FRQVQAEVPG 480
481 SPIFVMKLAR GARHLEVQLL ADQYGNAISL FGRDCSIQRR HQKIIEEAPA IVAQPEVFED 540
541 MEKAAVRLAK MVGYVSAGTV EYLYDPEGRY FFLELNPRLQ VEHPCTEMVA DVNLPAAQLQ 600
601 IGMGIPLYRL KDIRLLYGES PWGSSVIDFE NPPNKPRPSG HVIAARITSE NPDEGFKPSS 660
661 GTVQELNFRS SKNVWGYFSV AASGGLHEFA DSQFGHCFSW GENRQQAREN LVIALKELSI 720
721 RGDFRTTVEY LITLLETNRF LDNSIDTAWL DALIAERVQS EKPDILLGVM CGSLHIADRQ 780
781 ITESFSSFQT SLEKGQIQAA NTLTNVVDVE LINDGIRYKV QAAKSGANSY FLLMNSSFKE 840
841 IEVHRLSDGG LLISLEGASY TTYMKEEVDR YRIVIGNQTC VFEKENDPSL LRSPSAGKLI 900
901 NMIVEDGAHV SKGQAYAEIE VMKMVMTLTS QEAGTVTFVR RPGAVLDAGS LLGHLELDDP 960
961 SLVTKAQPFK GQFLQPENAP VPEKLNRVHN TYKSILENTL AGYCLPEPFN AQRLRDIIEK1020
1021 FMQSLRDPSL PLLELQEVIA SISGRIPISV EKKIRKLMTL YERNITSVLA QFPSQQIASV1080
1081 IDSHAATLQK RADRDVFFLT TQSIVQLVQR YRNGIRGRMK AAVHELLRQY YDVESQFQYG1140
1141 HYDKCVGLVR EHNKDDMQTV VNTIFSHSQV AKKNLLVTLL IDHLWANEPG LTDELANTLS1200
1201 ELTSLNRAEH SRVALRSRQV LIAAHQPAYE LRHNQMESIF LSAVDMYGHD FHPENLQRLI1260
1261 LSETSIFDIL HDFFYHSNRA VCNAALEVYV RRAYTSYELT CLQHLELSGG LPLVHFQFLL1320
1321 PTAHPNRLFS RMSSPDGLDQ AAAESLGNSF VRTGAIAAFD SFEHFEMYSD EILDLLEDFV1380
1381 SPAMVNAKVL EAVEAADSIS DSRHSTSINV SLSDPVTRAN AAEEAKSTEP IHIVSVAVRE1440
1441 TGELDDLQMA QIFGNYCQEH NEELFQRRIR RITFAALKKR QFPKFFTFRA RDKFTEDRIY1500
1501 RHLEPASAFH LELNRMKTYD LEALPTANQK MHLYLGKAKV SKGQEVTDYR FFIRSIIRHS1560
1561 DLITKEASFE YLQNEGERVL LEAMDELEVA FSHPHAKRTD CNHIFLNFVP TVIMDPAKIE1620
1621 ESVTKMIMRY GPRLWKLRVL QAELKMVIRQ SPQSPTQAVR LCIANDSGYF LDISMYTEQT1680
1681 EPETGIIKFK AYGEKQGSLH GHPISTPYMT KDFLQQKRFQ AQSNGTTYVY DVPDMFRQMT1740
1741 ERHWREFSKA RPTVDIRTPD KILIECKELV LEGDNLVEMQ RLPGENNCGM VAWRIVLATP1800
1801 EYPNGREIIV IANDLTYLIG SFGIKEDVLF AKASQLARQL KVPRIYISVN SGARIGLAEE1860
1861 VKAMFKIAWE DPEEPDKGFK YLYLSTEDYA QVANLNSVRA ILIEDEGEQR YKITDIIGKD1920
1921 DGLGVENLRY AGLIAGETSQ AYEEIVTIAM VTCRTIGIGS YVVRLGQRVI QIDNSHIILT1980
1981 GYAALNKLLG RKVYASNNQL GGTQIMFNNG VTHKTEAIDL DGVYTILDWL SYIPAYIGCD2040
2041 LPIVLPNDRI ERPVDFMPTK SPYDPRWMLG GRVNPVNAND WENGFFDRDS WSEIMASWAK2100
2101 TVVTGRARLG GVPVGVIAVE TRTVEVEMPA DPANLDSEAK TLQQAGQVWY PDSSYKTAQA2160
2161 IKDFGREELP LIVFANWRGF SGGMKDMYEQ IVKFGAYIVD GLREYKKPVL IYLPPNAELR2220
2221 GGAWAVLDSL INPRYMETYA DPEARGGVLE PEGIVEIKYK EKDLVKTIHR LDPTTIALKK2280
2281 ELDEANASGD KVRAAQVDEK IKARIAVLMH VYHTVAVHFA DLHDTPERML EKECISEIVP2340
2341 WRDSRRWLYW RLRRLLLEDA YIKKILRAQD NLSVGQAKQM LRRWLVEEKG ATEAYLWDKN2400
2401 EEMVSWYEEQ INAESIVSRN VNSVRRDAII STISKMLEDC PDVALDAVVG LCQGLTPVNR2460
2461 GVVVRTLAQM QLNEETSNSN QG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
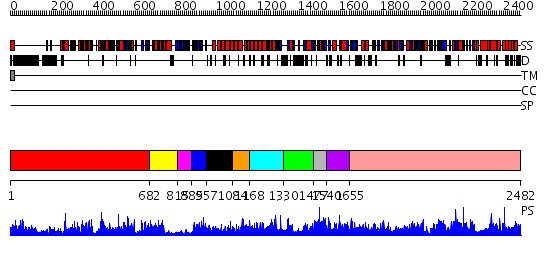
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..681] | 124.0 | Carbamoyl phosphate synthetase, large subunit connection domain; Carbamoyl phosphate synthetase, large subunit allosteric, C-terminal domain; Carbamoyl phosphate synthetase (CPS), large subunit PreATP-grasp domains; Carbamoyl phosphate synthetase (CPS), large subunit ATP-binding domains | |
| 2 | View Details | [682..814] | 147.0 | No description for 2hjwA was found. | |
| 3 | View Details | [815..884] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [885..956] | 17.045757 | No description for 2dn8A was found. | |
| 5 | View Details | [957..1083] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1084..1167] | 12.69897 | Dihydrolipoamide succinyltransferase | |
| 7 | View Details | [1168..1329] | 1.075966 | View MSA. No confident structure predictions are available. | |
| 8 | View Details | [1330..1476] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1477..1539] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1540..1654] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1655..2482] | 1000.0 | Acetyl-coenzyme A carboxylase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 11 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|