













| General Information: |
|
| Name(s) found: |
PEP4 /
YPL154C
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 405 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA fungal-type vacuole lumen [TAS fungal-type vacuole [TAS |
| Biological Process: |
cellular response to starvation
[IMP sporulation resulting in formation of a cellular spore [IMP vacuolar protein catabolic process [IMP microautophagy [IMP |
| Molecular Function: |
endopeptidase activity
[TAS saccharopepsin activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFSLKALLPL ALLLVSANQV AAKVHKAKIY KHELSDEMKE VTFEQHLAHL GQKYLTQFEK 60
61 ANPEVVFSRE HPFFTEGGHD VPLTNYLNAQ YYTDITLGTP PQNFKVILDT GSSNLWVPSN 120
121 ECGSLACFLH SKYDHEASSS YKANGTEFAI QYGTGSLEGY ISQDTLSIGD LTIPKQDFAE 180
181 ATSEPGLTFA FGKFDGILGL GYDTISVDKV VPPFYNAIQQ DLLDEKRFAF YLGDTSKDTE 240
241 NGGEATFGGI DESKFKGDIT WLPVRRKAYW EVKFEGIGLG DEYAELESHG AAIDTGTSLI 300
301 TLPSGLAEMI NAEIGAKKGW TGQYTLDCNT RDNLPDLIFN FNGYNFTIGP YDYTLEVSGS 360
361 CISAITPMDF PEPVGPLAIV GDAFLRKYYS IYDLGNNAVG LAKAI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
PEP4 RTN1 |
| View Details | Krogan NJ, et al. (2006) |
|
CRP1 IDS2 PDR1 PEP4 PRC1 RTN1 SIA1 |
| View Details | Gavin AC, et al. (2006) |

|
IDS2 PEP4 RTN1 YMR196W |
| View Details | Gavin AC, et al. (2002) |
|
ADH1 MIR1 PDC1 PEP4 RAD16 RIM1 TDH3 VMA4 VPS1 WRS1 YMR315W |
| View Details | Qiu et al. (2008) |
|
APE3 LAP4 PEP4 PRB1 PRC1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | experimental1 - mih1-1 | McCusker D, et al (2007) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi1 with gst from october 2005 | McCusker D, et al (2007) | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
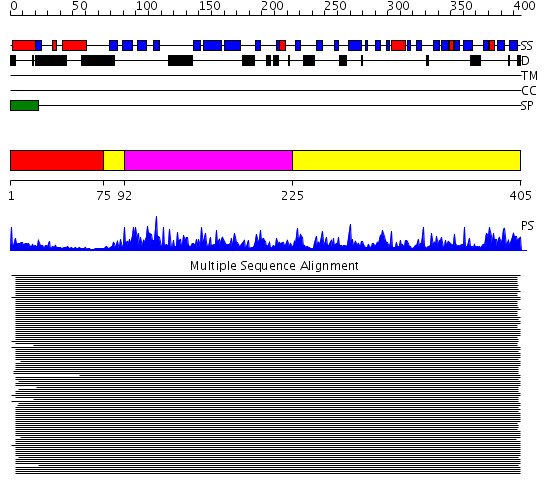
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..74] | N/A | Confident ab initio structure predictions are available. | |
| 2 | View Details | [75..91] [225..405] |
10123.0 | Acid protease | |
| 3 | View Details | [92..224] | 10123.0 | Acid protease |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|