













| General Information: |
|
| Name(s) found: |
ADE2 /
YOR128C
[SGD]
|
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 571 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
purine nucleotide biosynthetic process
[TAS]
'de novo' IMP biosynthetic process [TAS] purine base metabolic process [IMP |
| Molecular Function: |
phosphoribosylaminoimidazole carboxylase activity
[TAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDSRTVGILG GGQLGRMIVE AANRLNIKTV ILDAENSPAK QISNSNDHVN GSFSNPLDIE 60
61 KLAEKCDVLT IEIEHVDVPT LKNLQVKHPK LKIYPSPETI RLIQDKYIQK EHLIKNGIAV 120
121 TQSVPVEQAS ETSLLNVGRD LGFPFVLKSR TLAYDGRGNF VVKNKEMIPE ALEVLKDRPL 180
181 YAEKWAPFTK ELAVMIVRSV NGLVFSYPIV ETIHKDNICD LCYAPARVPD SVQLKAKLLA 240
241 ENAIKSFPGC GIFGVEMFYL ETGELLINEI APRPHNSGHY TIDACVTSQF EAHLRSILDL 300
301 PMPKNFTSFS TITTNAIMLN VLGDKHTKDK ELETCERALA TPGSSVYLYG KESRPNRKVG 360
361 HINIIASSMA ECEQRLNYIT GRTDIPIKIS VAQKLDLEAM VKPLVGIIMG SDSDLPVMSA 420
421 ACAVLKDFGV PFEVTIVSAH RTPHRMSAYA ISASKRGIKT IIAGAGGAAH LPGMVAAMTP 480
481 LPVIGVPVKG SCLDGVDSLH SIVQMPRGVP VATVAINNST NAALLAVRLL GAYDSSYTTK 540
541 MEQFLLKQEE EVLVKAQKLE TVGYEAYLEN K |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
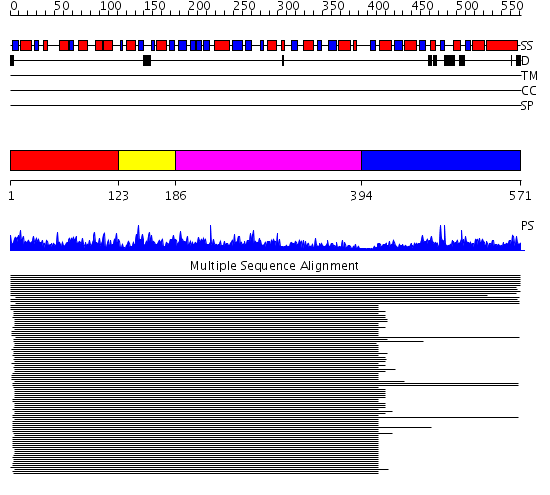
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..122] | 342.917458 | N5-carboxyaminoimidazole ribonucleotide synthetase PurK (AIRC), C-domain; N5-carboxyaminoimidazole ribonucleotide synthetase PurK (AIRC), N-domain; N5-carboxyaminoimidazole ribonucleotide synthetase PurK (AIRC), domain 2 | |
| 2 | View Details | [123..185] | 342.917458 | N5-carboxyaminoimidazole ribonucleotide synthetase PurK (AIRC), C-domain; N5-carboxyaminoimidazole ribonucleotide synthetase PurK (AIRC), N-domain; N5-carboxyaminoimidazole ribonucleotide synthetase PurK (AIRC), domain 2 | |
| 3 | View Details | [186..393] | 342.917458 | N5-carboxyaminoimidazole ribonucleotide synthetase PurK (AIRC), C-domain; N5-carboxyaminoimidazole ribonucleotide synthetase PurK (AIRC), N-domain; N5-carboxyaminoimidazole ribonucleotide synthetase PurK (AIRC), domain 2 | |
| 4 | View Details | [394..571] | 304.045757 | N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.75 |
Source: Reynolds et al. (2008)