













| General Information: |
|
| Name(s) found: |
NTG2 /
YOL043C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 380 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
base-excision repair
[IDA base-excision repair, AP site formation [IDA |
| Molecular Function: |
oxidized pyrimidine base lesion DNA N-glycosylase activity
[IDA DNA-(apurinic or apyrimidinic site) lyase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MREESRSRKR KHIPVDIEEV EVRSKYFKKN ERTVELVKEN KINKDLQNYG GVNIDWIKAL 60
61 KPIEYFEWIE SRTCDDPRTW GRPITKEEMI NDSGAKVPES FLPIYNRVRL MRSKVKTPVD 120
121 AMGCSMIPVL VSNKCGIPSE KVDPKNFRLQ FLIGTMLSAQ TRDERMAQAA LNITEYCLNT 180
181 LKIAEGITLD GLLKIDEPVL ANLIRCVSFY TRKANFIKRT AQLLVDNFDS DIPYDIEGIL 240
241 SLPGVGPKMG YLTLQKGWGL IAGICVDVHV HRLCKMWNWV DPIKCKTAEH TRKELQVWLP 300
301 HSLWYEINTV LVGFGQLICM ARGKRCDLCL ANDVCNARNE KLIESSKFHQ LEDKEDIEKV 360
361 YSHWLDTVTN GITTERHKKK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
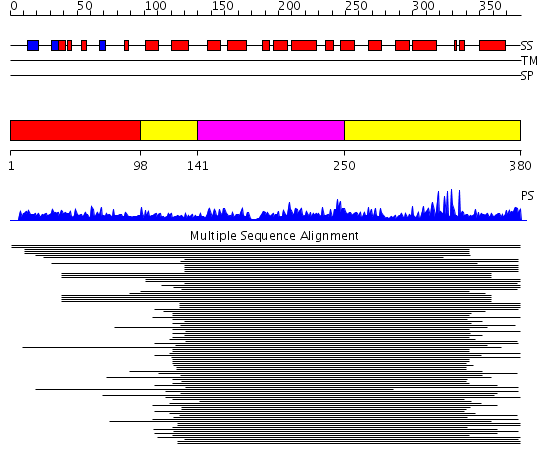
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..97] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [98..140] [250..380] |
170.927757 | Endonuclease III | |
| 3 | View Details | [141..249] | 170.927757 | Endonuclease III |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)