













| General Information: |
|
| Name(s) found: |
MAS1 /
YLR163C
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 462 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA mitochondrial processing peptidase complex [IDA |
| Biological Process: |
protein processing involved in protein targeting to mitochondrion
[IDA |
| Molecular Function: |
mitochondrial processing peptidase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFSRTASKFR NTRRLLSTIS SQIPGTRTSK LPNGLTIATE YIPNTSSATV GIFVDAGSRA 60
61 ENVKNNGTAH FLEHLAFKGT QNRSQQGIEL EIENIGSHLN AYTSRENTVY YAKSLQEDIP 120
121 KAVDILSDIL TKSVLDNSAI ERERDVIIRE SEEVDKMYDE VVFDHLHEIT YKDQPLGRTI 180
181 LGPIKNIKSI TRTDLKDYIT KNYKGDRMVL AGAGAVDHEK LVQYAQKYFG HVPKSESPVP 240
241 LGSPRGPLPV FCRGERFIKE NTLPTTHIAI ALEGVSWSAP DYFVALATQA IVGNWDRAIG 300
301 TGTNSPSPLA VAASQNGSLA NSYMSFSTSY ADSGLWGMYI VTDSNEHNVQ LIVNEILKEW 360
361 KRIKSGKISD AEVNRAKAQL KAALLLSLDG STAIVEDIGR QVVTTGKRLS PEEVFEQVDK 420
421 ITKDDIIMWA NYRLQNKPVS MVALGNTSTV PNVSYIEEKL NQ |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
MAS1 MAS2 |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
MAS1 MAS2 |
| View Details | Krogan NJ, et al. (2006) |
|
ATG2 MAS1 MAS2 |
| View Details | Gavin AC, et al. (2006) |

|
MAS1 MAS2 |
| View Details | Gavin AC, et al. (2002) |
|
DIS3 ISW1 MAS1 MAS2 RRP4 SMC4 |
| View Details | Ho Y, et al. (2002) |
|
MAS1 MAS2 MSH1 RNR3 |
| View Details | Qiu et al. (2008) |

|
MAS1 MAS2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
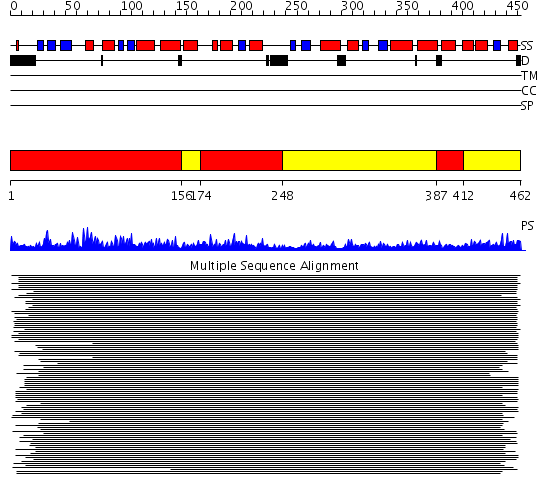
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..155] [174..247] [387..411] |
10127.0 | Mitochondrial processing peptidase (MPP) beta chain | |
| 2 | View Details | [156..173] [248..386] [412..462] |
10127.0 | Mitochondrial processing peptidase (MPP) beta chain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|