













| General Information: |
|
| Name(s) found: |
GCD6 /
YDR211W
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 712 amino acids |
Gene Ontology: |
|
| Cellular Component: |
guanyl-nucleotide exchange factor complex
[IDA eukaryotic translation initiation factor 2B complex [IDA |
| Biological Process: |
regulation of translational initiation
[IDA |
| Molecular Function: |
translation initiation factor activity
[IGI guanyl-nucleotide exchange factor activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAGKKGQKKS GLGNHGKNSD MDVEDRLQAV VLTDSYETRF MPLTAVKPRC LLPLANVPLI 60
61 EYTLEFLAKA GVHEVFLICS SHANQINDYI ENSKWNLPWS PFKITTIMSP EARCTGDVMR 120
121 DLDNRGIITG DFILVSGDVL TNIDFSKMLE FHKKMHLQDK DHISTMCLSK ASTYPKTRTI 180
181 EPAAFVLDKS TSRCIYYQDL PLPSSREKTS IQIDPELLDN VDEFVIRNDL IDCRIDICTS 240
241 HVPLIFQENF DYQSLRTDFV KGVISSDILG KHIYAYLTDE YAVRVESWQT YDTISQDFLG 300
301 RWCYPLVLDS NIQDDQTYSY ESRHIYKEKD VVLAQSCKIG KCTAIGSGTK IGEGTKIENS 360
361 VIGRNCQIGE NIRIKNSFIW DDCIIGNNSI IDHSLIASNA TLGSNVRLND GCIIGFNVKI 420
421 DDNMDLDRNT KISASPLKNA GSRMYDNESN EQFDQDLDDQ TLAVSIVGDK GVGYIYESEV 480
481 SDDEDSSTEA CKEINTLSNQ LDELYLSDDS ISSATKKTKK RRTMSVNSIY TDREEIDSEF 540
541 EDEDFEKEGI ATVERAMENN HDLDTALLEL NTLRMSMNVT YHEVRIATIT ALLRRVYHFI 600
601 ATQTLGPKDA VVKVFNQWGL LFKRQAFDEE EYIDLMNIIM EKIVEQSFDK PDLILFSALV 660
661 SLYDNDIIEE DVIYKWWDNV STDPRYDEVK KLTVKWVEWL QNADEESSSE EE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
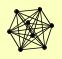
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
GCD1 GCD11 GCD2 GCD6 GCD7 GCN3 SUI2 SUI3 |
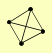
| View Details | Hazbun TR, et al. (2003) |
|
GCD6 NOP9 NSR1 SNU13 |
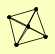
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
GCD1 GCD2 GCD6 GCD7 GCN3 |
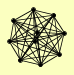
| View Details | Krogan NJ, et al. (2006) |
|
ADE1 GCD1 GCD11 GCD2 GCD6 GCD7 GCN3 MRF1 MRH4 SUI2 |
| View Details | Gavin AC, et al. (2006) |
|
GCD1 GCD11 GCD2 GCD6 GCD7 GCN3 |
| View Details | Gavin AC, et al. (2002) |
|
ASC1 CCT4 CHC1 CKA1 COG8 FKS1 GCD1 GCD11 GCD2 GCD6 GCD7 GCN3 GUT1 IST1 NAT1 NEM1 PDC1 PRO1 RRP40 RRP46 SCP160 SUI2 SUI3 SWI3 XKS1 |
| View Details | Ho Y, et al. (2002) |
|
CDC33 FAA4 FAL1 FET3 GCD1 GCD11 GCD2 GCD6 GCD7 GCN3 LOS1 MET18 NMD5 PRO3 SCW4 SUI2 SUI3 TIF1, TIF2 TIF4631 TIF4632 TUP1 VAC8 VMA8 |
| View Details | Qiu et al. (2008) |
|
GCD1 GCD2 GCD6 GCD7 GCN3 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
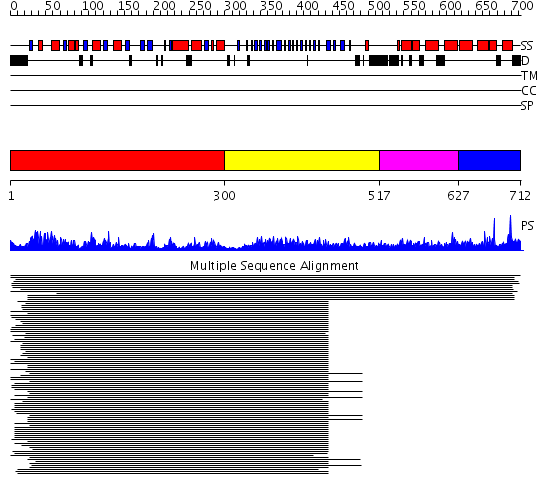
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..299] | 116.917458 | N-acetylglucosamine 1-phosphate uridyltransferase GlmU, C-terminal domain; N-acetylglucosamine 1-phosphate uridyltransferase GlmU, N-terminal domain | |
| 2 | View Details | [300..516] | 116.917458 | N-acetylglucosamine 1-phosphate uridyltransferase GlmU, C-terminal domain; N-acetylglucosamine 1-phosphate uridyltransferase GlmU, N-terminal domain | |
| 3 | View Details | [517..626] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [627..712] | 27.60206 | eIF4-gamma/eIF5/eIF2-epsilon No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)