













| General Information: |
|
| Name(s) found: |
SOR2 /
YDL246C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 357 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
hexose metabolic process
[RCA |
| Molecular Function: |
oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQNSNPAVV LEKVGDIAIE QRPIPTIKDP HYVKLAIKAT GICGSDIHYY RSGGIGKYIL 60
61 KAPMVLGHES SGQVVEVGDA VTRVKVGDRV AIEPGVPSRY SDETKEGSYN LCPHMAFAAT 120
121 PPIDGTLVKY YLSPEDFLVK LPEGVSYEEG ACVEPLSVGV HSNKLAGVRF GTKVVVFGAG 180
181 PVGLLTGAVA RAFGATDVIF VDVFDNKLQR AKDFGATNTF NSSQFSTDKA QDLADGVQKL 240
241 LGGNHADVVF ECSGADVCID AAVKTTKVGG TMVQVGMGKN YTNFPIAEVS GKEMKLIGCF 300
301 RYSFGDYRDA VNLVATGKVN VKPLITHKFK FEDAAKAYDY NIAHGGEVVK TIIFGPE |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
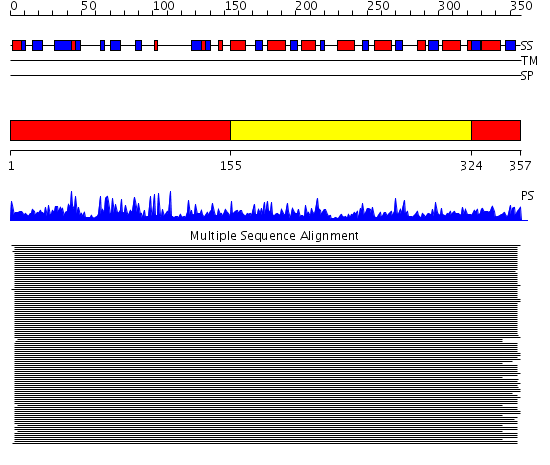
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..154] [324..357] |
851.39794 | Ketose reductase (sorbitol dehydrogenase) | |
| 2 | View Details | [155..323] | 851.39794 | Ketose reductase (sorbitol dehydrogenase) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.86 |
Source: Reynolds et al. (2008)