













| General Information: |
|
| Name(s) found: |
DOT1 /
YDR440W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 582 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
DNA damage checkpoint
[IGI recombinational repair [IGI regulation of meiosis [IMP histone methylation [IDA nucleotide-excision repair [IGI chromatin silencing at telomere [IMP postreplication repair [IGI |
| Molecular Function: |
protein-lysine N-methyltransferase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGGQESISNN NSDSFIMSSP NLDSQESSIS PIDEKKGTDM QTKSLSSYSK GTLLSKQVQN 60
61 LLEEANKYDP IYGSSLPRGF LRDRNTKGKD NGLVPLVEKV IPPIHKKTNN RNTRKKSSTT 120
121 TKKDVKKPKA AKVKGKNGRT NHKHTPISKQ EIDTAREKKP LKKGRANKKN DRDSPSSTFV 180
181 DWNGPCLRLQ YPLFDIEYLR SHEIYSGTPI QSISLRTNSP QPTSLTSDND TSSVTTAKLQ 240
241 SILFSNYMEE YKVDFKRSTA IYNPMSEIGK LIEYSCLVFL PSPYAEQLKE TILPDLNASF 300
301 DNSDTKGFVN AINLYNKMIR EIPRQRIIDH LETIDKIPRS FIHDFLHIVY TRSIHPQANK 360
361 LKHYKAFSNY VYGELLPNFL SDVYQQCQLK KGDTFMDLGS GVGNCVVQAA LECGCALSFG 420
421 CEIMDDASDL TILQYEELKK RCKLYGMRLN NVEFSLKKSF VDNNRVAELI PQCDVILVNN 480
481 FLFDEDLNKK VEKILQTAKV GCKIISLKSL RSLTYQINFY NVENIFNRLK VQRYDLKEDS 540
541 VSWTHSGGEY YISTVMEDVD ESLFSPAARG RRNRGTPVKY TR |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
DBP8 DOT1 IMH1 TRM1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
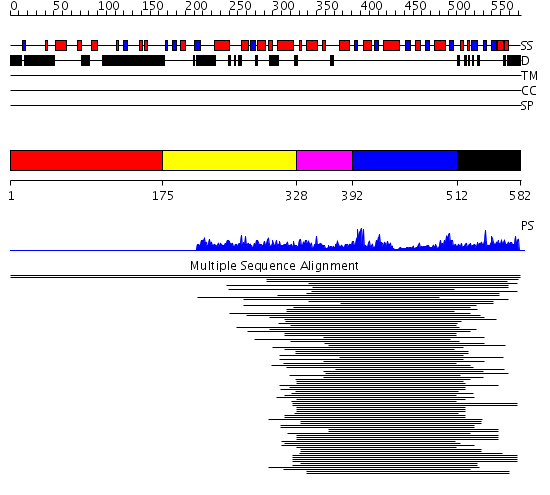
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..174] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [175..327] | 1.143996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [328..391] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [392..511] | 680.211549 | No description for 1m0rA_ was found. | |
| 5 | View Details | [512..582] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)