













| General Information: |
|
| Name(s) found: |
LBX1_MOUSE
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Mus musculus |
| Length: | 282 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA]
transcription factor complex [IDA] |
| Biological Process: |
transcription
[IEA]
multicellular organismal development [IEA] negative regulation of cell proliferation [IMP] nervous system development [IEA] negative regulation of neuron differentiation [IMP] regulation of transcription, DNA-dependent [IDA] regulation of transcription from RNA polymerase II promoter involved in spinal cord association neuron specification [IMP] regulation of transcription [IEA] muscle organ development [IEA] neuron fate determination [IGI][IMP] neuron fate commitment [IMP] cell differentiation [IEA] heart looping [IMP] |
| Molecular Function: |
DNA binding
[IEA]
transcription factor activity [IDA] protein binding [IPI] transcription regulator activity [IEA] sequence-specific DNA binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTSKEDGKAA PGEERRRSPL DHLPPPANSN KPLTPFSIED ILNKPSVRRS YSLCGAAHLL 60
61 AAADKHAPGG LPLAGRALLS QTSPLCALEE LASKTFKGLE VSVLQAAEGR DGMTIFGQRQ 120
121 TPKKRRKSRT AFTNHQIYEL EKRFLYQKYL SPADRDQIAQ QLGLTNAQVI TWFQNRRAKL 180
181 KRDLEEMKAD VESAKKLGPS GQMDIVALAE LEQNSEASGG GGGGGCGRAK SRPGSPALPP 240
241 GAPQAPGGGP LQLSPASPLT DQRASSQDCS EDEEDEEIDV DD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
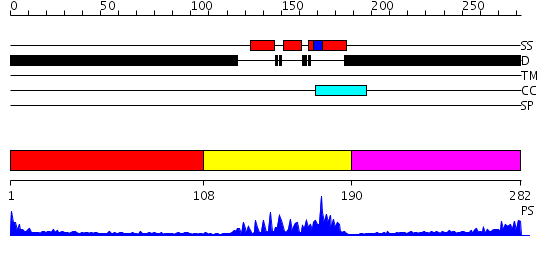
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..107] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [108..189] | 24.69897 | Crystal Structure of HoxA9 and Pbx1 homeodomains bound to DNA | |
| 3 | View Details | [190..282] | 2.288996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)